当前位置:
X-MOL 学术
›
J. Cell. Biochem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Construction and evaluation of a prognosis lncRNA model for hepatocellular carcinoma
Journal of Cellular Biochemistry ( IF 3.0 ) Pub Date : 2020-04-29 , DOI: 10.1002/jcb.29608 Fulei Li 1 , Lu Bai 1 , Shasha Li 1 , Yanling Chen 1 , Xiaofei Xue 2 , Zujiang Yu 1
Journal of Cellular Biochemistry ( IF 3.0 ) Pub Date : 2020-04-29 , DOI: 10.1002/jcb.29608 Fulei Li 1 , Lu Bai 1 , Shasha Li 1 , Yanling Chen 1 , Xiaofei Xue 2 , Zujiang Yu 1
Affiliation
|
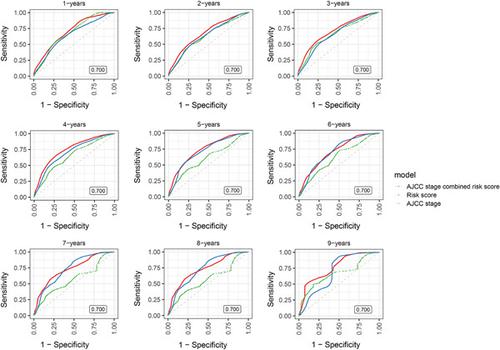
Current studies indicate that long non-coding RNA (lncRNA) is often abnormally expressed in hepatocellular carcinoma (HCC). We intend to generate a multi-lncRNA signal to improve the prognosis of HCC. By analyzing 12 pairs of HCC and adjacent normal mucosal tissues, 3900 differentially expressed lncrnas were identified as candidate biomarkers for the prognosis of HCC. Then, the 12-lncrna signature was constructed using the LASSO Cox regression method and verified in the TCGA training dataset. Finally, we established a novel 12-lncrna signature that was significantly associated with overall survival (OS) in the training data set. With the use of 12-lncrna markers, patients in the training cohort were divided into high-risk and low-risk groups with significant OV differences (P < .0001). Similar results were consistent in the TCGA verification dataset (P = .046). Multivariate Cox model was used to analyze and construct the risk scores of selected key lncRNA and AJCC stages. The results showed that, compared with AJCC stages, lncRNA-based risk scores were another important factor affecting the OS of patients. We found that risk scores based on lncRNA have a stronger prediction ability than the AJCC stage alone on 4-year OS. For 4-year survival rates, prediction combined with the lncRNA risk score and AJCC stage, model effectiveness (sensitivity and specificity) has reached to 0.750. To further explore the biological processes involved in prognostic lncRNA, all HCC samples in TCGA are divided into two groups according to the median lncRNA risk score, and analyzed the gene enrichment of high expression genes and low expression genes in KEGG data using goana in limma. The results suggest that the genes associated with tumor pathways, such as PI3K-Akt and ECM-receptor interaction, are highly expressed in the high risk group.
中文翻译:

肝细胞癌lncRNA预后模型的构建与评估
目前的研究表明,长链非编码RNA(lncRNA)在肝细胞癌(HCC)中经常异常表达。我们打算产生多lncRNA信号来改善HCC的预后。通过分析12对HCC和癌旁正常粘膜组织,鉴定出3900个差异表达的lncrnas作为HCC预后的候选生物标志物。然后,使用LASSO Cox回归方法构建12-lncrna签名,并在TCGA训练数据集中进行验证。最后,我们建立了一种新颖的 12-lncrna 特征,该特征与训练数据集中的总生存期 (OS) 显着相关。通过使用 12-lncrna 标记,将训练队列中的患者分为高风险组和低风险组,OV 差异显着 ( P < .0001)。类似的结果在 TCGA 验证数据集中是一致的 ( P = .046)。使用多变量 Cox 模型分析和构建所选关键 lncRNA 和 AJCC 阶段的风险评分。结果显示,与AJCC分期相比,基于lncRNA的风险评分是影响患者OS的另一个重要因素。我们发现基于 lncRNA 的风险评分比单独的 AJCC 阶段对 4 年 OS 具有更强的预测能力。对于4年生存率,结合lncRNA风险评分和AJCC分期进行预测,模型有效性(敏感性和特异性)已达到0.750。为了进一步探讨lncRNA参与预后的生物学过程,TCGA中的所有HCC样本根据中位lncRNA风险评分分为两组,并使用goana in limma分析KEGG数据中高表达基因和低表达基因的基因富集度。 结果表明,与肿瘤通路相关的基因,如 PI3K-Akt 和 ECM-受体相互作用,在高危人群中高度表达。
更新日期:2020-04-29
中文翻译:

肝细胞癌lncRNA预后模型的构建与评估
目前的研究表明,长链非编码RNA(lncRNA)在肝细胞癌(HCC)中经常异常表达。我们打算产生多lncRNA信号来改善HCC的预后。通过分析12对HCC和癌旁正常粘膜组织,鉴定出3900个差异表达的lncrnas作为HCC预后的候选生物标志物。然后,使用LASSO Cox回归方法构建12-lncrna签名,并在TCGA训练数据集中进行验证。最后,我们建立了一种新颖的 12-lncrna 特征,该特征与训练数据集中的总生存期 (OS) 显着相关。通过使用 12-lncrna 标记,将训练队列中的患者分为高风险组和低风险组,OV 差异显着 ( P < .0001)。类似的结果在 TCGA 验证数据集中是一致的 ( P = .046)。使用多变量 Cox 模型分析和构建所选关键 lncRNA 和 AJCC 阶段的风险评分。结果显示,与AJCC分期相比,基于lncRNA的风险评分是影响患者OS的另一个重要因素。我们发现基于 lncRNA 的风险评分比单独的 AJCC 阶段对 4 年 OS 具有更强的预测能力。对于4年生存率,结合lncRNA风险评分和AJCC分期进行预测,模型有效性(敏感性和特异性)已达到0.750。为了进一步探讨lncRNA参与预后的生物学过程,TCGA中的所有HCC样本根据中位lncRNA风险评分分为两组,并使用goana in limma分析KEGG数据中高表达基因和低表达基因的基因富集度。 结果表明,与肿瘤通路相关的基因,如 PI3K-Akt 和 ECM-受体相互作用,在高危人群中高度表达。











































 京公网安备 11010802027423号
京公网安备 11010802027423号