Structure ( IF 4.4 ) Pub Date : 2020-05-05 , DOI: 10.1016/j.str.2020.04.003 Aron Broom 1 , Kyle Trainor 1 , Zachary Jacobi 1 , Elizabeth M Meiering 1
|
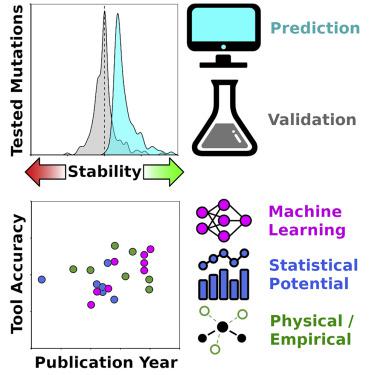
Accurate modeling of the effects of mutations on protein stability is central to understanding and controlling proteins in myriad natural and applied contexts. Here, we reveal through rigorous quantitative analysis that stability prediction tools often favor mutations that increase stability at the expense of solubility. Moreover, while these tools may accurately identify strongly destabilizing mutations, the experimental effect of mutations predicted to stabilize is actually near neutral on average. The commonly used “classification accuracy” metric obscures this reality; accordingly, we recommend performance measures, such as the Matthews correlation coefficient (MCC). We demonstrate that an absurdly simple machine-learning algorithm—a neural network of just two neurons—unexpectedly achieves high classification accuracy, but its inadequacies are revealed by a low MCC. Despite the above limitations, making multiple mutations markedly improves the prospects for achieving a stabilization target, and modest improvements in the precision of future tools may yield disproportionate gains.
中文翻译:

蛋白质稳定性的计算模型:定量分析揭示了普遍存在的问题的解决方案。
准确模拟突变对蛋白质稳定性的影响,对于了解和控制多种自然环境和应用环境中的蛋白质至关重要。在这里,我们通过严格的定量分析揭示了稳定性预测工具通常偏爱以增加溶解度为代价而增加稳定性的突变。此外,尽管这些工具可以准确地识别出严重破坏稳定性的突变,但预测稳定的突变的实验效果实际上平均接近中性。常用的“分类准确性”度量标准掩盖了这一现实;因此,我们建议采用性能指标,例如马修斯相关系数(MCC)。我们证明了一种荒谬的简单机器学习算法(只有两个神经元的神经网络)意外地实现了很高的分类精度,但MCC偏低表明了它的不足。尽管存在上述限制,但进行多个突变可以显着改善实现稳定目标的前景,并且对未来工具的精度进行适度的改进可能会产生不成比例的收益。











































 京公网安备 11010802027423号
京公网安备 11010802027423号