当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Deep complex convolutional network for fast reconstruction of 3D late gadolinium enhancement cardiac MRI.
NMR in Biomedicine ( IF 2.7 ) Pub Date : 2020-04-30 , DOI: 10.1002/nbm.4312 Hossam El-Rewaidy 1, 2 , Ulf Neisius 1 , Jennifer Mancio 1 , Selcuk Kucukseymen 1 , Jennifer Rodriguez 1 , Amanda Paskavitz 1 , Bjoern Menze 2 , Reza Nezafat 1
NMR in Biomedicine ( IF 2.7 ) Pub Date : 2020-04-30 , DOI: 10.1002/nbm.4312 Hossam El-Rewaidy 1, 2 , Ulf Neisius 1 , Jennifer Mancio 1 , Selcuk Kucukseymen 1 , Jennifer Rodriguez 1 , Amanda Paskavitz 1 , Bjoern Menze 2 , Reza Nezafat 1
Affiliation
|
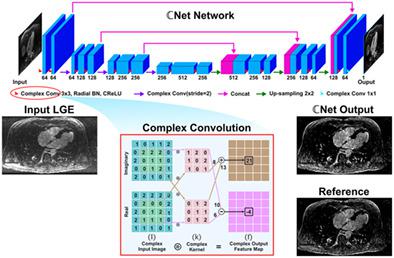
Several deep‐learning models have been proposed to shorten MRI scan time. Prior deep‐learning models that utilize real‐valued kernels have limited capability to learn rich representations of complex MRI data. In this work, we utilize a complex‐valued convolutional network (ℂ Net) for fast reconstruction of highly under‐sampled MRI data and evaluate its ability to rapidly reconstruct 3D late gadolinium enhancement (LGE) data. ℂ Net preserves the complex nature and optimal combination of real and imaginary components of MRI data throughout the reconstruction process by utilizing complex‐valued convolution, novel radial batch normalization, and complex activation function layers in a U‐Net architecture. A prospectively under‐sampled 3D LGE cardiac MRI dataset of 219 patients (17 003 images) at acceleration rates R = 3 through R = 5 was used to evaluate ℂ Net. The dataset was further retrospectively under‐sampled to a maximum of R = 8 to simulate higher acceleration rates. We created three reconstructions of the 3D LGE dataset using (1) ℂ Net, (2) a compressed‐sensing‐based low‐dimensional‐structure self‐learning and thresholding algorithm (LOST), and (3) a real‐valued U‐Net (realNet) with the same number of parameters as ℂ Net. LOST‐reconstructed data were considered the reference for training and evaluation of all models. The reconstructed images were quantitatively evaluated using mean‐squared error (MSE) and the structural similarity index measure (SSIM), and subjectively evaluated by three independent readers. Quantitatively, ℂ Net‐reconstructed images had significantly improved MSE and SSIM values compared with realNet (MSE, 0.077 versus 0.091; SSIM, 0.876 versus 0.733, respectively; p < 0.01). Subjective quality assessment showed that ℂ Net‐reconstructed image quality was similar to that of compressed sensing and significantly better than that of realNet. ℂ Net reconstruction was also more than 300 times faster than compressed sensing. Retrospective under‐sampled images demonstrate the potential of ℂ Net at higher acceleration rates. ℂ Net enables fast reconstruction of highly accelerated 3D MRI with superior performance to real‐valued networks, and achieves faster reconstruction than compressed sensing.
中文翻译:

用于快速重建 3D 晚期钆增强心脏 MRI 的深度复杂卷积网络。
已经提出了几种深度学习模型来缩短 MRI 扫描时间。先前使用实值核的深度学习模型在学习复杂 MRI 数据的丰富表示方面的能力有限。在这项工作中,我们利用复值卷积网络 ( ℂ Net ) 来快速重建高度欠采样的 MRI 数据,并评估其快速重建 3D 晚期钆增强 (LGE) 数据的能力。ℂ Net通过在 U-Net 架构中利用复值卷积、新颖的径向批量归一化和复杂的激活函数层,在整个重建过程中保留了 MRI 数据的实部和虚部的复杂性质和最佳组合。一个前瞻性的219 名患者(17 003 幅图像)的欠采样 3D LGE 心脏 MRI 数据集以R = 3 到R = 5 的加速度用于评估ℂ Net。数据集进一步回溯性欠采样到最大R = 8,以模拟更高的加速度。我们使用 (1) ℂ Net、(2) 基于压缩感知的低维结构自学习和阈值算法 (LOST) 以及 (3) 实值 U-具有与ℂ Net相同数量参数的 Net (realNet). LOST 重建的数据被认为是所有模型训练和评估的参考。使用均方误差 (MSE) 和结构相似性指数度量 (SSIM) 对重建的图像进行定量评估,并由三个独立的读者进行主观评估。从数量上讲,与 realNet 相比,ℂ Net重建图像的 MSE 和 SSIM 值显着提高(MSE,分别为 0.077 对 0.091;SSIM,分别为 0.876 对 0.733;p < 0.01)。主观质量评估表明,ℂ Net重建的图像质量与压缩感知相似,明显优于 realNet。ℂ净重建速度也比压缩感知快 300 多倍。回顾性欠采样图像证明了ℂ Net在更高的加速度下的潜力。ℂ Net能够快速重建高加速 3D MRI,其性能优于实值网络,并实现比压缩感知更快的重建。
更新日期:2020-04-30
中文翻译:

用于快速重建 3D 晚期钆增强心脏 MRI 的深度复杂卷积网络。
已经提出了几种深度学习模型来缩短 MRI 扫描时间。先前使用实值核的深度学习模型在学习复杂 MRI 数据的丰富表示方面的能力有限。在这项工作中,我们利用复值卷积网络 ( ℂ Net ) 来快速重建高度欠采样的 MRI 数据,并评估其快速重建 3D 晚期钆增强 (LGE) 数据的能力。ℂ Net通过在 U-Net 架构中利用复值卷积、新颖的径向批量归一化和复杂的激活函数层,在整个重建过程中保留了 MRI 数据的实部和虚部的复杂性质和最佳组合。一个前瞻性的219 名患者(17 003 幅图像)的欠采样 3D LGE 心脏 MRI 数据集以R = 3 到R = 5 的加速度用于评估ℂ Net。数据集进一步回溯性欠采样到最大R = 8,以模拟更高的加速度。我们使用 (1) ℂ Net、(2) 基于压缩感知的低维结构自学习和阈值算法 (LOST) 以及 (3) 实值 U-具有与ℂ Net相同数量参数的 Net (realNet). LOST 重建的数据被认为是所有模型训练和评估的参考。使用均方误差 (MSE) 和结构相似性指数度量 (SSIM) 对重建的图像进行定量评估,并由三个独立的读者进行主观评估。从数量上讲,与 realNet 相比,ℂ Net重建图像的 MSE 和 SSIM 值显着提高(MSE,分别为 0.077 对 0.091;SSIM,分别为 0.876 对 0.733;p < 0.01)。主观质量评估表明,ℂ Net重建的图像质量与压缩感知相似,明显优于 realNet。ℂ净重建速度也比压缩感知快 300 多倍。回顾性欠采样图像证明了ℂ Net在更高的加速度下的潜力。ℂ Net能够快速重建高加速 3D MRI,其性能优于实值网络,并实现比压缩感知更快的重建。











































 京公网安备 11010802027423号
京公网安备 11010802027423号