IRBM ( IF 5.6 ) Pub Date : 2020-01-03 , DOI: 10.1016/j.irbm.2019.12.004 R. Touati , I. Messaoudi , A.E. Oueslati , Z. Lachiri , M. Kharrat
|
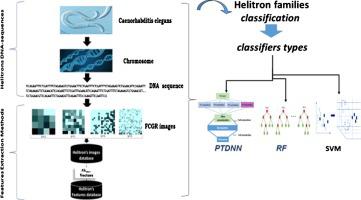
Helitrons, eukaryotic transposable elements (TEs) transposed by rolling-circle mechanism, have been found in various species with highly variable copy numbers and sometimes with a large portion of their genomes. The impact of helitrons sequences in the genome is to frequently capture host genes during their transposition. Since their discovery, 18 years ago, by computational analysis of whole genome sequences of Arabidopsis thaliana plant and Caenorhabditis elegans (C. elegans) nematode, the identification and classification of these mobile genetic elements remain a challenge due to the fact that the wide majority of their families are non-autonomous. In C. elegans genome, DNA helitrons sequences possess great variability in terms of length that varies between 11 and 8965 base pairs (bps) from one sequence to another. In this work, we develop a new method to predict helitrons DNA-sequences, which is particularly based on Frequency Chaos Game Representation (FCGR) DNA-images. Thus, we introduce an automatic system in order to classify helitrons families in C. elegans genome, based on a combination between machine learning approaches and features extracted from DNA-sequences. Consequently, the new set of helitrons features (the FCGR images and K-mers) are extracted from DNA sequences. These helitrons features consist of the frequency apparition number of K nucleotides pairs (Tandem Repeat) in the DNA sequences. Indeed, three different classifiers are used for the classification of all existing helitrons families. The results have shown potential global score equal to 72.7% due to FCGR images which constitute helitrons features and the pre-trained neural network as a classifier. The two other classifiers demonstrate that their efficiency reaches 68.7% for Support Vector Machine (SVM) and 91.45% for Random Forest (RF) algorithms using the K-mers features corresponding to the genomic sequences.
中文翻译:

使用DNA图像序列和机器学习方法的新的类内Helitrons分类
直升飞机是通过滚环机制转位的真核转座因子(TEs),已在各种物种中发现了拷贝数高度可变且有时其基因组很大一部分的物种。Helitrons序列在基因组中的影响是在转座过程中频繁捕获宿主基因。自18年前发现以来,通过计算分析拟南芥植物和秀丽隐杆线虫(C. elegans)线虫的全基因组序列,由于大多数他们的家庭是非自治的。在秀丽隐杆线虫中在基因组中,DNA Helitrons序列在长度上具有很大的可变性,从一个序列到另一个序列在11到8965个碱基对(bps)之间变化。在这项工作中,我们开发了一种预测高等电子人DNA序列的新方法,该方法特别基于频率混沌博弈表示(FCGR)DNA图像。因此,我们引入了一个自动系统,以便对秀丽隐杆线虫中的高等电子族进行分类基因组,基于机器学习方法和从DNA序列中提取的特征之间的组合。因此,从DNA序列中提取了一组新的高电子特征(FCGR图像和K-mers)。这些高电子特征包括DNA序列中K个核苷酸对的频率显性数(串联重复)。确实,使用三种不同的分类器对所有现有的直升机场系列进行分类。结果显示,由于FCGR图像(构成了直升机的特征)和预先训练的神经网络作为分类器,潜在的全局评分等于72.7%。另外两个分类器证明,使用对应于基因组序列的K-mers特征,对支持向量机(SVM)的效率达到68.7%,对随机森林(RF)算法的效率达到91.45%。











































 京公网安备 11010802027423号
京公网安备 11010802027423号