当前位置:
X-MOL 学术
›
Mol. Omics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
ProteoViz: a tool for the analysis and interactive visualization of phosphoproteomics data.
Molecular Omics ( IF 3.0 ) Pub Date : 2020-04-29 , DOI: 10.1039/c9mo00149b Aaron J Storey 1 , Kevin S Naceanceno , Renny S Lan , Charity L Washam , Lisa M Orr , Samuel G Mackintosh , Alan J Tackett , Rick D Edmondson , Zhengyu Wang , Hong-Yu Li , Brendan Frett , Samantha Kendrick , Stephanie D Byrum
Molecular Omics ( IF 3.0 ) Pub Date : 2020-04-29 , DOI: 10.1039/c9mo00149b Aaron J Storey 1 , Kevin S Naceanceno , Renny S Lan , Charity L Washam , Lisa M Orr , Samuel G Mackintosh , Alan J Tackett , Rick D Edmondson , Zhengyu Wang , Hong-Yu Li , Brendan Frett , Samantha Kendrick , Stephanie D Byrum
Affiliation
|
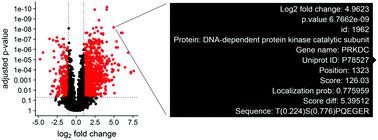
Quantitative proteomics generates large datasets with increasing depth and quantitative information. With the advance of mass spectrometry and increasingly larger data sets, streamlined methodologies and tools for analysis and visualization of phosphoproteomics are needed both at the protein and modified peptide levels. To assist in addressing this need, we developed ProteoViz, which includes a set of R scripts that perform normalization and differential expression analysis of both the proteins and enriched phosphorylated peptides, and identify sequence motifs, kinases, and gene set enrichment pathways. The tool generates interactive visualization plots that allow users to interact with the phosphoproteomics results and quickly identify proteins and phosphorylated peptides of interest for their biological study. The tool also links significant phosphosites with sequence motifs and pathways that will help explain the experimental conditions and guide future experiments. Here, we present the workflow and demonstrate its functionality by analyzing a phosphoproteomic data set from two lymphoma cell lines treated with kinase inhibitors. The scripts and data are freely available at https://github.com/ByrumLab/ProteoViz and via the ProteomeXchange with identifier PXD015606.
中文翻译:

ProteoViz:一种用于磷酸化蛋白质组学数据分析和交互式可视化的工具。
定量蛋白质组学生成具有不断增加的深度和定量信息的大型数据集。随着质谱技术的进步和越来越大的数据集,在蛋白质和修饰肽水平上都需要用于磷酸化蛋白质组学分析和可视化的简化方法和工具。为了帮助满足这一需求,我们开发了 ProteoViz,其中包括一组 R 脚本,可对蛋白质和富集的磷酸化肽进行标准化和差异表达分析,并识别序列基序、激酶和基因集富集途径。该工具生成交互式可视化图,允许用户与磷酸蛋白质组学结果进行交互,并快速识别其生物学研究中感兴趣的蛋白质和磷酸化肽。该工具还将重要的磷酸位点与序列基序和通路联系起来,这将有助于解释实验条件并指导未来的实验。在这里,我们通过分析来自用激酶抑制剂处理的两个淋巴瘤细胞系的磷酸化蛋白质组数据集来介绍工作流程并证明其功能。脚本和数据可在 https://github.com/ByrumLab/ProteoViz 和通过带有标识符 PXD015606 的 ProteomeXchange。
更新日期:2020-04-29
中文翻译:

ProteoViz:一种用于磷酸化蛋白质组学数据分析和交互式可视化的工具。
定量蛋白质组学生成具有不断增加的深度和定量信息的大型数据集。随着质谱技术的进步和越来越大的数据集,在蛋白质和修饰肽水平上都需要用于磷酸化蛋白质组学分析和可视化的简化方法和工具。为了帮助满足这一需求,我们开发了 ProteoViz,其中包括一组 R 脚本,可对蛋白质和富集的磷酸化肽进行标准化和差异表达分析,并识别序列基序、激酶和基因集富集途径。该工具生成交互式可视化图,允许用户与磷酸蛋白质组学结果进行交互,并快速识别其生物学研究中感兴趣的蛋白质和磷酸化肽。该工具还将重要的磷酸位点与序列基序和通路联系起来,这将有助于解释实验条件并指导未来的实验。在这里,我们通过分析来自用激酶抑制剂处理的两个淋巴瘤细胞系的磷酸化蛋白质组数据集来介绍工作流程并证明其功能。脚本和数据可在 https://github.com/ByrumLab/ProteoViz 和通过带有标识符 PXD015606 的 ProteomeXchange。











































 京公网安备 11010802027423号
京公网安备 11010802027423号