Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure ( IF 4.4 ) Pub Date : 2020-04-28 , DOI: 10.1016/j.str.2020.04.002 Mathieu Lussier-Price 1 , Xavier H Mascle 1 , Laurent Cappadocia 1 , Rui Kamada 2 , Kazuyasu Sakaguchi 2 , Haytham M Wahba 3 , James G Omichinski 1
Structure ( IF 4.4 ) Pub Date : 2020-04-28 , DOI: 10.1016/j.str.2020.04.002 Mathieu Lussier-Price 1 , Xavier H Mascle 1 , Laurent Cappadocia 1 , Rui Kamada 2 , Kazuyasu Sakaguchi 2 , Haytham M Wahba 3 , James G Omichinski 1
Affiliation
|
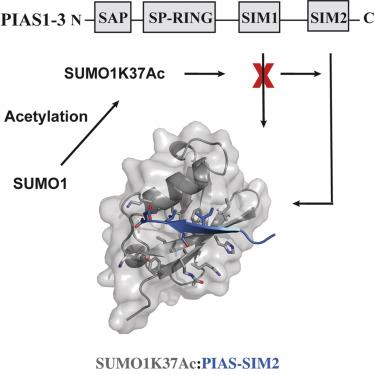
The human PIAS proteins are small ubiquitin-like modifier (SUMO) E3 ligases that participate in important cellular functions. Several of these functions depend on a conserved SUMO-interacting motif (SIM) located in the central region of all PIAS proteins (SIM1). Recently, it was determined that Siz2, a yeast homolog of PIAS proteins, possesses a second SIM at its C terminus (SIM2). Sequence alignment indicates that a SIM2 is also present in PIAS1-3, but not PIAS4. Using biochemical and structural studies, we demonstrate PIAS-SIM2 binds to SUMO1, but that phosphorylation of the PIAS-SIM2 or acetylation of SUMO1 alter this interaction in a manner distinct from what is observed for the PIAS-SIM1. We also show that the PIAS-SIM2 plays a key role in formation of a UBC9-PIAS1-SUMO1 complex. These results provide insights into how post-translational modifications selectively regulate the specificity of multiple SIMs found in the PIAS proteins by exploiting the plasticity built into the SUMO-SIM binding interface.
中文翻译:

选择性PIAS家族蛋白中存在的C端SUMO交互基序的表征。
人PIAS蛋白是参与重要细胞功能的小型泛素样修饰剂(SUMO)E3连接酶。其中一些功能取决于位于所有PIAS蛋白(SIM1)中心区域的保守SUMO相互作用基序(SIM)。最近,已确定PIAS蛋白的酵母同源物Siz2在其C末端(SIM2)具有第二个SIM。序列比对表明在PIAS1-3中也存在SIM2,但在PIAS4中不存在。使用生化和结构研究,我们证明PIAS-SIM2与SUMO1结合,但是PIAS-SIM2的磷酸化或SUMO1的乙酰化以不同于PIAS-SIM1的方式改变了这种相互作用。我们还显示,PIAS-SIM2在UBC9-PIAS1-SUMO1复合体的形成中起关键作用。
更新日期:2020-04-28
中文翻译:

选择性PIAS家族蛋白中存在的C端SUMO交互基序的表征。
人PIAS蛋白是参与重要细胞功能的小型泛素样修饰剂(SUMO)E3连接酶。其中一些功能取决于位于所有PIAS蛋白(SIM1)中心区域的保守SUMO相互作用基序(SIM)。最近,已确定PIAS蛋白的酵母同源物Siz2在其C末端(SIM2)具有第二个SIM。序列比对表明在PIAS1-3中也存在SIM2,但在PIAS4中不存在。使用生化和结构研究,我们证明PIAS-SIM2与SUMO1结合,但是PIAS-SIM2的磷酸化或SUMO1的乙酰化以不同于PIAS-SIM1的方式改变了这种相互作用。我们还显示,PIAS-SIM2在UBC9-PIAS1-SUMO1复合体的形成中起关键作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号