当前位置:
X-MOL 学术
›
Hum. Mutat.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
EFTUD2 missense variants disrupt protein function and splicing in mandibulofacial dysostosis Guion-Almeida type.
Human Mutation ( IF 3.3 ) Pub Date : 2020-04-25 , DOI: 10.1002/humu.24027 Huw B Thomas 1 , Katherine A Wood 1 , Weronika A Buczek 1 , Christopher T Gordon 2, 3 , Véronique Pingault 2, 3, 4 , Tania Attié-Bitach 3, 4, 5 , Kathryn E Hentges 1 , Vinod C Varghese 6 , Jeanne Amiel 2, 3, 4 , William G Newman 1, 7 , Raymond T O'Keefe 1
Human Mutation ( IF 3.3 ) Pub Date : 2020-04-25 , DOI: 10.1002/humu.24027 Huw B Thomas 1 , Katherine A Wood 1 , Weronika A Buczek 1 , Christopher T Gordon 2, 3 , Véronique Pingault 2, 3, 4 , Tania Attié-Bitach 3, 4, 5 , Kathryn E Hentges 1 , Vinod C Varghese 6 , Jeanne Amiel 2, 3, 4 , William G Newman 1, 7 , Raymond T O'Keefe 1
Affiliation
|
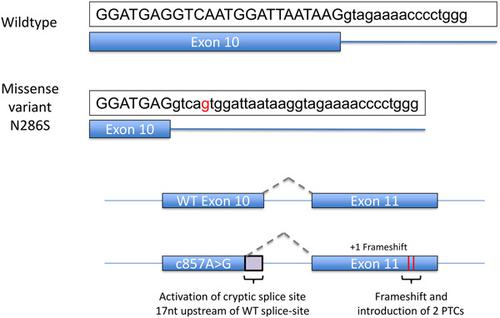
Pathogenic variants in the core spliceosome U5 small nuclear ribonucleoprotein gene EFTUD2/SNU114 cause the craniofacial disorder mandibulofacial dysostosis Guion‐Almeida type (MFDGA). MFDGA‐associated variants in EFTUD2 comprise large deletions encompassing EFTUD2 , intragenic deletions and single nucleotide truncating or missense variants. These variants are predicted to result in haploinsufficiency by loss‐of‐function of the variant allele. While the contribution of deletions within EFTUD2 to allele loss‐of‐function are self‐evident, the mechanisms by which missense variants are disease‐causing have not been characterized functionally. Combining bioinformatics software prediction, yeast functional growth assays, and a minigene (MG) splicing assay, we have characterized how MFDGA missense variants result in EFTUD2 loss‐of‐function. Only four of 19 assessed missense variants cause EFTUD2 loss‐of‐function through altered protein function when modeled in yeast. Of the remaining 15 missense variants, five altered the normal splicing pattern of EFTUD2 pre‐messenger RNA predominantly through exon skipping or cryptic splice site activation, leading to the introduction of a premature termination codon. Comparison of bioinformatic predictors for each missense variant revealed a disparity amongst different software packages and, in many cases, an inability to correctly predict changes in splicing subsequently determined by MG interrogation. This study highlights the need for laboratory‐based validation of bioinformatic predictions for EFTUD2 missense variants.
中文翻译:

EFTUD2 错义变异破坏了下颌面骨发育不良 Guion-Almeida 型的蛋白质功能和剪接。
核心剪接体 U5 小核核糖核蛋白基因EFTUD2/SNU114中的致病变异导致颅面疾病下颌面骨发育不良 Guion-Almeida 型 (MFDGA)。在MFDGA相关变体EFTUD2包括大的缺失包含EFTUD2,基因内缺失和单核苷酸截取或错义变异体。预计这些变异会因变异等位基因的功能丧失而导致单倍体不足。虽然EFTUD2中删除的贡献对于等位基因功能丧失是不言而喻的,错义变异导致疾病的机制尚未在功能上表征。结合生物信息学软件预测、酵母功能生长测定和小基因 (MG) 剪接测定,我们表征了 MFDGA 错义变异如何导致EFTUD2功能丧失。在酵母中建模时,19 个评估的错义变异中只有 4 个通过改变蛋白质功能导致EFTUD2功能丧失。在剩下的 15 个错义变体中,有五个改变了EFTUD2的正常剪接模式前信使 RNA 主要通过外显子跳跃或神秘剪接位点激活,导致引入提前终止密码子。每个错义变体的生物信息学预测因子的比较揭示了不同软件包之间的差异,并且在许多情况下,无法正确预测随后由 MG 询问确定的剪接变化。这项研究强调了对EFTUD2错义变异的生物信息学预测进行实验室验证的必要性。
更新日期:2020-04-25
中文翻译:

EFTUD2 错义变异破坏了下颌面骨发育不良 Guion-Almeida 型的蛋白质功能和剪接。
核心剪接体 U5 小核核糖核蛋白基因EFTUD2/SNU114中的致病变异导致颅面疾病下颌面骨发育不良 Guion-Almeida 型 (MFDGA)。在MFDGA相关变体EFTUD2包括大的缺失包含EFTUD2,基因内缺失和单核苷酸截取或错义变异体。预计这些变异会因变异等位基因的功能丧失而导致单倍体不足。虽然EFTUD2中删除的贡献对于等位基因功能丧失是不言而喻的,错义变异导致疾病的机制尚未在功能上表征。结合生物信息学软件预测、酵母功能生长测定和小基因 (MG) 剪接测定,我们表征了 MFDGA 错义变异如何导致EFTUD2功能丧失。在酵母中建模时,19 个评估的错义变异中只有 4 个通过改变蛋白质功能导致EFTUD2功能丧失。在剩下的 15 个错义变体中,有五个改变了EFTUD2的正常剪接模式前信使 RNA 主要通过外显子跳跃或神秘剪接位点激活,导致引入提前终止密码子。每个错义变体的生物信息学预测因子的比较揭示了不同软件包之间的差异,并且在许多情况下,无法正确预测随后由 MG 询问确定的剪接变化。这项研究强调了对EFTUD2错义变异的生物信息学预测进行实验室验证的必要性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号