当前位置:
X-MOL 学术
›
Prog. Nucl. Magn. Reson. Spectrosc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Automated assignment of methyl NMR spectra from large proteins
Progress in Nuclear Magnetic Resonance Spectroscopy ( IF 7.3 ) Pub Date : 2020-06-01 , DOI: 10.1016/j.pnmrs.2020.04.001 Iva Pritišanac 1 , T Reid Alderson 2 , Peter Güntert 3
Progress in Nuclear Magnetic Resonance Spectroscopy ( IF 7.3 ) Pub Date : 2020-06-01 , DOI: 10.1016/j.pnmrs.2020.04.001 Iva Pritišanac 1 , T Reid Alderson 2 , Peter Güntert 3
Affiliation
|
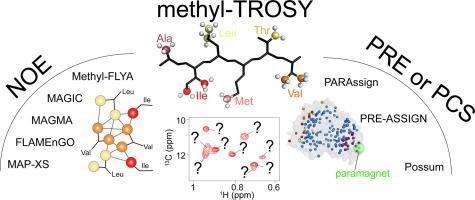
As structural biology trends towards larger and more complex biomolecular targets, a detailed understanding of their interactions and underlying structures and dynamics is required. The development of methyl-TROSY has enabled NMR spectroscopy to provide atomic-resolution insight into the mechanisms of large molecular assemblies in solution. However, the applicability of methyl-TROSY has been hindered by the laborious and time-consuming resonance assignment process, typically performed with domain fragmentation, site-directed mutagenesis, and analysis of NOE data in the context of a crystal structure. In response, several structure-based automatic methyl assignment strategies have been developed over the past decade. Here, we present a comprehensive analysis of all available methods and compare their input data requirements, algorithmic strategies, and reported performance. In general, the methods fall into two categories: those that primarily rely on inter-methyl NOEs, and those that utilize methyl PRE- and PCS-based restraints. We discuss their advantages and limitations, and highlight the potential benefits from standardizing and combining different methods.
中文翻译:

自动分配大蛋白的甲基 NMR 谱
随着结构生物学趋向于更大和更复杂的生物分子目标,需要详细了解它们的相互作用以及潜在的结构和动力学。甲基 TROSY 的发展使核磁共振光谱能够提供对溶液中大分子组装机制的原子分辨率洞察。然而,甲基-TROSY 的适用性受到费力和耗时的共振分配过程的阻碍,通常通过结构域碎片、定点诱变和在晶体结构的背景下分析 NOE 数据来进行。作为回应,在过去十年中开发了几种基于结构的自动甲基分配策略。在这里,我们对所有可用方法进行了全面分析,并比较了它们的输入数据要求、算法策略、并报告了性能。一般而言,这些方法分为两类:主要依赖于甲基间 NOE 的方法,以及利用基于甲基 PRE 和 PCS 限制的方法。我们讨论了它们的优点和局限性,并强调了标准化和组合不同方法的潜在好处。
更新日期:2020-06-01
中文翻译:

自动分配大蛋白的甲基 NMR 谱
随着结构生物学趋向于更大和更复杂的生物分子目标,需要详细了解它们的相互作用以及潜在的结构和动力学。甲基 TROSY 的发展使核磁共振光谱能够提供对溶液中大分子组装机制的原子分辨率洞察。然而,甲基-TROSY 的适用性受到费力和耗时的共振分配过程的阻碍,通常通过结构域碎片、定点诱变和在晶体结构的背景下分析 NOE 数据来进行。作为回应,在过去十年中开发了几种基于结构的自动甲基分配策略。在这里,我们对所有可用方法进行了全面分析,并比较了它们的输入数据要求、算法策略、并报告了性能。一般而言,这些方法分为两类:主要依赖于甲基间 NOE 的方法,以及利用基于甲基 PRE 和 PCS 限制的方法。我们讨论了它们的优点和局限性,并强调了标准化和组合不同方法的潜在好处。











































 京公网安备 11010802027423号
京公网安备 11010802027423号