Structure ( IF 4.4 ) Pub Date : 2020-04-21 , DOI: 10.1016/j.str.2020.03.011 Maybelle Kho Go 1 , Li Na Zhao 2 , Bo Xue 1 , Shreyas Supekar 2 , Robert C Robinson 3 , Hao Fan 2 , Wen Shan Yew 1
|
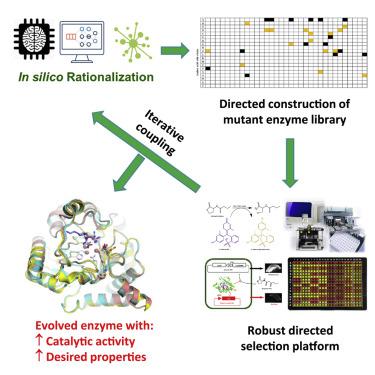
In this work, we present a generalizable directed computational evolution protocol to effectively reduce the sequence space to be explored in rational enzyme design. The protocol involves in silico mutation modeling and substrate docking to rapidly identify mutagenesis hotspots that may enhance an enzyme's substrate binding and overall catalysis. By applying this protocol to a quorum-quenching Geobacillus kaustophilus lactonase, GKL, we generated 1,881 single mutants and docked high-energy intermediates of nine acyl homoserine lactones onto them. We found that Phe28 and Tyr99 were two hotspots that produced most of the predicted top 20 mutants. Of the 180 enzyme-substrate combinations (top 20 mutants × 9 substrates), 51 (28%) exhibited enhanced substrate binding and 22 (12%) had better overall activity when compared with wild-type GKL. X-ray crystallographic studies of Y99C and Y99P provided rationalized explanations for the enhancement in enzyme function and corroborated the utility of the protocol.
中文翻译:

酰胺水解酶超家族的猝灭乳糖酶的定向计算进化。
在这项工作中,我们提出了一种可概括的定向计算进化协议,以有效减少合理的酶设计中要探索的序列空间。该协议涉及计算机突变建模和底物对接,以快速识别诱变热点,该热点可能会增强酶的底物结合和整体催化作用。通过将此协议应用于仲裁Quaubacillus kaustophilus内酯酶GKL,我们产生了1,881个单一突变体,并将9个酰基高丝氨酸内酯的高能中间体对接在其上。我们发现Phe28和Tyr99是两个热点,产生了大多数预测的前20个突变体。与野生型GKL相比,在180种酶-底物组合中(前20个突变体×9种底物),有51种(28%)显示出增强的底物结合,而22种(12%)的总体活性更好。Y99C和Y99P的X射线晶体学研究为增强酶功能提供了合理的解释,并证实了该方案的实用性。










































 京公网安备 11010802027423号
京公网安备 11010802027423号