当前位置:
X-MOL 学术
›
PLOS Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Resident microbial communities inhibit growth and antibiotic-resistance evolution of Escherichia coli in human gut microbiome samples.
PLOS Biology ( IF 7.8 ) Pub Date : 2020-04-20 , DOI: 10.1371/journal.pbio.3000465 Michael Baumgartner 1 , Florian Bayer 2 , Katia R Pfrunder-Cardozo 1 , Angus Buckling 2 , Alex R Hall 1
PLOS Biology ( IF 7.8 ) Pub Date : 2020-04-20 , DOI: 10.1371/journal.pbio.3000465 Michael Baumgartner 1 , Florian Bayer 2 , Katia R Pfrunder-Cardozo 1 , Angus Buckling 2 , Alex R Hall 1
Affiliation
|
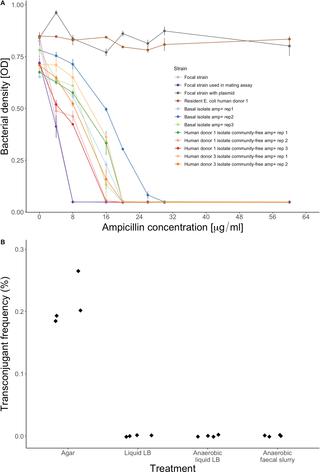
Countering the rise of antibiotic-resistant pathogens requires improved understanding of how resistance emerges and spreads in individual species, which are often embedded in complex microbial communities such as the human gut microbiome. Interactions with other microorganisms in such communities might suppress growth and resistance evolution of individual species (e.g., via resource competition) but could also potentially accelerate resistance evolution via horizontal transfer of resistance genes. It remains unclear how these different effects balance out, partly because it is difficult to observe them directly. Here, we used a gut microcosm approach to quantify the effect of three human gut microbiome communities on growth and resistance evolution of a focal strain of Escherichia coli. We found the resident microbial communities not only suppressed growth and colonisation by focal E. coli but also prevented it from evolving antibiotic resistance upon exposure to a beta-lactam antibiotic. With samples from all three human donors, our focal E. coli strain only evolved antibiotic resistance in the absence of the resident microbial community, even though we found resistance genes, including a highly effective resistance plasmid, in resident microbial communities. We identified physical constraints on plasmid transfer that can explain why our focal strain failed to acquire some of these beneficial resistance genes, and we found some chromosomal resistance mutations were only beneficial in the absence of the resident microbiota. This suggests, depending on in situ gene transfer dynamics, interactions with resident microbiota can inhibit antibiotic-resistance evolution of individual species.
中文翻译:

常驻微生物群落抑制人类肠道微生物样本中大肠杆菌的生长和抗生素耐药性进化。
应对抗生素耐药性病原体的增加需要更好地了解耐药性如何在单个物种中出现和传播,这些物种通常嵌入在复杂的微生物群落中,例如人类肠道微生物组。与此类群落中其他微生物的相互作用可能会抑制单个物种的生长和抗性进化(例如,通过资源竞争),但也可能通过抗性基因的水平转移加速抗性进化。目前尚不清楚这些不同的影响如何平衡,部分原因是很难直接观察它们。在这里,我们使用肠道微观方法来量化三种人类肠道微生物群落对大肠杆菌重点菌株的生长和耐药性进化的影响。我们发现,常驻微生物群落不仅抑制了焦点大肠杆菌的生长和定植,而且还阻止其在接触β-内酰胺抗生素后产生抗生素耐药性。对于来自所有三个人类捐赠者的样本,我们的焦点大肠杆菌菌株仅在没有常驻微生物群落的情况下进化出抗生素耐药性,尽管我们在常驻微生物群落中发现了耐药基因,包括高效的耐药质粒。我们确定了质粒转移的物理限制,这可以解释为什么我们的焦点菌株未能获得其中一些有益的抗性基因,并且我们发现一些染色体抗性突变仅在缺乏常驻微生物群的情况下才有益。这表明,根据原位基因转移动力学,与常驻微生物群的相互作用可以抑制单个物种的抗生素抗性进化。
更新日期:2020-04-20
中文翻译:

常驻微生物群落抑制人类肠道微生物样本中大肠杆菌的生长和抗生素耐药性进化。
应对抗生素耐药性病原体的增加需要更好地了解耐药性如何在单个物种中出现和传播,这些物种通常嵌入在复杂的微生物群落中,例如人类肠道微生物组。与此类群落中其他微生物的相互作用可能会抑制单个物种的生长和抗性进化(例如,通过资源竞争),但也可能通过抗性基因的水平转移加速抗性进化。目前尚不清楚这些不同的影响如何平衡,部分原因是很难直接观察它们。在这里,我们使用肠道微观方法来量化三种人类肠道微生物群落对大肠杆菌重点菌株的生长和耐药性进化的影响。我们发现,常驻微生物群落不仅抑制了焦点大肠杆菌的生长和定植,而且还阻止其在接触β-内酰胺抗生素后产生抗生素耐药性。对于来自所有三个人类捐赠者的样本,我们的焦点大肠杆菌菌株仅在没有常驻微生物群落的情况下进化出抗生素耐药性,尽管我们在常驻微生物群落中发现了耐药基因,包括高效的耐药质粒。我们确定了质粒转移的物理限制,这可以解释为什么我们的焦点菌株未能获得其中一些有益的抗性基因,并且我们发现一些染色体抗性突变仅在缺乏常驻微生物群的情况下才有益。这表明,根据原位基因转移动力学,与常驻微生物群的相互作用可以抑制单个物种的抗生素抗性进化。









































 京公网安备 11010802027423号
京公网安备 11010802027423号