当前位置:
X-MOL 学术
›
Chem. Res. Toxicol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Comparison of the Base Excision and Direct Reversal Repair Pathways for Correcting 1,N6-Ethenoadenine in Strongly Positioned Nucleosome Core Particles.
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2020-04-15 , DOI: 10.1021/acs.chemrestox.0c00089 Paul J Caffrey 1 , Raadhika Kher 1 , Ke Bian 2 , Deyu Li 2 , Sarah Delaney 1
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2020-04-15 , DOI: 10.1021/acs.chemrestox.0c00089 Paul J Caffrey 1 , Raadhika Kher 1 , Ke Bian 2 , Deyu Li 2 , Sarah Delaney 1
Affiliation
|
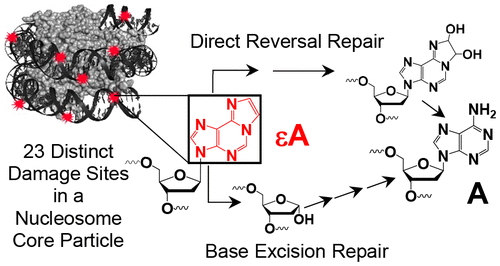
1,N6-ethenoadenine (εA) is a mutagenic lesion and biomarker observed in numerous cancerous tissues. Two pathways are responsible for its repair: base excision repair (BER) and direct reversal repair (DRR). Alkyladenine DNA glycosylase (AAG) is the primary enzyme that excises εA in BER, generating stable intermediates that are processed by downstream enzymes. For DRR, the Fe(II)/α-ketoglutarate-dependent ALKBH2 enzyme repairs εA by direct conversion of εA to A. While the molecular mechanism of each enzyme is well understood on unpackaged duplex DNA, less is known about their actions on packaged DNA. The nucleosome core particle (NCP) forms the minimal packaging unit of DNA in eukaryotic organisms and is composed of 145–147 base pairs wrapped around a core of eight histone proteins. In this work, we investigated the activity of AAG and ALKBH2 on εA lesions globally distributed at positions throughout a strongly positioned NCP. Overall, we examined the repair of εA at 23 unique locations in packaged DNA. We observed a strong correlation between rotational positioning of εA and AAG activity but not ALKBH2 activity. ALKBH2 was more effective than AAG at repairing occluded εA lesions, but only AAG was capable of full repair of any εA in the NCP. However, notable exceptions to these trends were observed, highlighting the complexity of the NCP as a substrate for DNA repair. Modeling of binding of the repair enzymes to NCPs revealed that some of these observations can be explained by steric interference caused by DNA packaging. Specifically, interactions between ALKBH2 and the histone proteins obstruct binding to DNA, which leads to diminished activity. Taken together, these results support in vivo observations of alkylation damage profiles and contribute to our understanding of mutational hotspots.
中文翻译:

碱基切除和直接逆向修复途径在强定位核小体核心颗粒中校正1,N6-乙腺嘌呤的比较。
1,N 6乙炔腺嘌呤(εA)是在许多癌性组织中观察到的诱变性病变和生物标志物。修复的途径有两种:碱基切除修复(BER)和直接逆转修复(DRR)。烷基腺嘌呤DNA糖基化酶(AAG)是在BER中切除εA的主要酶,可生成稳定的中间体,并由下游酶加工。对于DRR,依赖于Fe(II)/α-酮戊二酸的ALKBH2酶通过将εA直接转化为A来修复εA。虽然每种酶的分子机理在未包装的双链DNA上已广为人知,但对其在包装DNA上的作用了解较少。 。核小体核心颗粒(NCP)构成了真核生物中DNA的最小包装单位,由围绕八个组蛋白的核心包裹的145-147个碱基对组成。在这项工作中 我们研究了AAG和ALKBH2对εA病灶的活性,该病灶分布在整个强力NCP位置。总的来说,我们检查了包装DNA中23个独特位置上εA的修复。我们观察到εA的旋转定位和AAG活性之间有很强的相关性,但ALKBH2活性却没有。ALKBH2在修复被闭塞的εA病变方面比AAG更为有效,但只有AAG能够完全修复NCP中的任何εA。但是,观察到这些趋势的显着例外,突显了NCP作为DNA修复底物的复杂性。修复酶与NCP结合的模型表明,其中一些观察结果可以解释为DNA包装引起的空间干扰。具体而言,ALKBH2与组蛋白之间的相互作用阻碍了与DNA的结合,这导致活动减少。综上所述,这些结果支持体内对烷基化损伤分布的观察,有助于我们对突变热点的了解。
更新日期:2020-04-15
中文翻译:

碱基切除和直接逆向修复途径在强定位核小体核心颗粒中校正1,N6-乙腺嘌呤的比较。
1,N 6乙炔腺嘌呤(εA)是在许多癌性组织中观察到的诱变性病变和生物标志物。修复的途径有两种:碱基切除修复(BER)和直接逆转修复(DRR)。烷基腺嘌呤DNA糖基化酶(AAG)是在BER中切除εA的主要酶,可生成稳定的中间体,并由下游酶加工。对于DRR,依赖于Fe(II)/α-酮戊二酸的ALKBH2酶通过将εA直接转化为A来修复εA。虽然每种酶的分子机理在未包装的双链DNA上已广为人知,但对其在包装DNA上的作用了解较少。 。核小体核心颗粒(NCP)构成了真核生物中DNA的最小包装单位,由围绕八个组蛋白的核心包裹的145-147个碱基对组成。在这项工作中 我们研究了AAG和ALKBH2对εA病灶的活性,该病灶分布在整个强力NCP位置。总的来说,我们检查了包装DNA中23个独特位置上εA的修复。我们观察到εA的旋转定位和AAG活性之间有很强的相关性,但ALKBH2活性却没有。ALKBH2在修复被闭塞的εA病变方面比AAG更为有效,但只有AAG能够完全修复NCP中的任何εA。但是,观察到这些趋势的显着例外,突显了NCP作为DNA修复底物的复杂性。修复酶与NCP结合的模型表明,其中一些观察结果可以解释为DNA包装引起的空间干扰。具体而言,ALKBH2与组蛋白之间的相互作用阻碍了与DNA的结合,这导致活动减少。综上所述,这些结果支持体内对烷基化损伤分布的观察,有助于我们对突变热点的了解。










































 京公网安备 11010802027423号
京公网安备 11010802027423号