当前位置:
X-MOL 学术
›
Nat. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Genetically engineered control of phenotypic structure in microbial colonies.
Nature Microbiology ( IF 20.5 ) Pub Date : 2020-04-13 , DOI: 10.1038/s41564-020-0686-0 Philip Bittihn 1, 2, 3 , Andriy Didovyk 1, 4 , Lev S Tsimring 1, 2 , Jeff Hasty 1, 2, 5, 6
Nature Microbiology ( IF 20.5 ) Pub Date : 2020-04-13 , DOI: 10.1038/s41564-020-0686-0 Philip Bittihn 1, 2, 3 , Andriy Didovyk 1, 4 , Lev S Tsimring 1, 2 , Jeff Hasty 1, 2, 5, 6
Affiliation
|
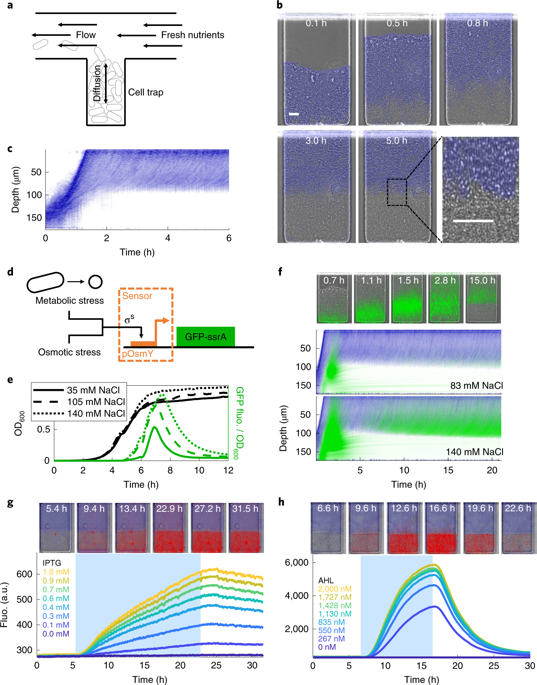
Rapid advances in cellular engineering1,2 have positioned synthetic biology to address therapeutic3,4 and industrial5 problems, but a substantial obstacle is the myriad of unanticipated cellular responses in heterogeneous real-world environments such as the gut6,7, solid tumours8,9, bioreactors10 or soil11. Complex interactions between the environment and cells often arise through non-uniform nutrient availability, which generates bidirectional coupling as cells both adjust to and modify their local environment through phenotypic differentiation12,13. Although synthetic spatial gene expression patterns14-17 have been explored under homogeneous conditions, the mutual interaction of gene circuits, growth phenotype and the environment remains a challenge. Here, we design gene circuits that sense and control phenotypic structure in microcolonies containing both growing and dormant bacteria. We implement structure modulation by coupling different downstream modules to a tunable sensor that leverages Escherichia coli's stress response and is activated on growth arrest. One is an actuator module that slows growth and thereby alters nutrient gradients. Environmental feedback in this circuit generates robust cycling between growth and dormancy in the interior of the colony, as predicted by a spatiotemporal computational model. We also use the sensor to drive an inducible gating module for selective gene expression in non-dividing cells, which allows us to radically alter population structure by eliminating the dormant phenotype with a 'stress-gated lysis circuit'. Our results establish a strategy to leverage and control microbial colony structure for synthetic biology applications in complex environments.
中文翻译:

微生物菌落表型结构的基因工程控制。
细胞工程 1,2 的快速发展使合成生物学能够解决治疗 3,4 和工业 5 问题,但一个重大障碍是在异质的现实世界环境(如肠道 6,7、实体瘤 8、9、生物反应器 10)中出现无数意料之外的细胞反应或土壤 11。环境和细胞之间的复杂相互作用通常是通过不均匀的营养可用性产生的,当细胞通过表型分化12、13 适应和改变其局部环境时,这会产生双向耦合。尽管已在均质条件下探索了合成空间基因表达模式14-17,但基因回路、生长表型和环境的相互作用仍然是一个挑战。这里,我们设计的基因电路可以感知和控制包含生长和休眠细菌的微菌落中的表型结构。我们通过将不同的下游模块耦合到一个可调节的传感器来实现结构调制,该传感器利用大肠杆菌的应激反应并在生长停滞时被激活。一种是减慢生长并因此改变营养梯度的执行器模块。正如时空计算模型所预测的那样,该电路中的环境反馈会在菌落内部产生生长和休眠之间的稳健循环。我们还使用传感器驱动一个可诱导的门控模块,用于在非分裂细胞中进行选择性基因表达,这使我们能够通过使用“压力门控裂解电路”消除休眠表型来从根本上改变种群结构。
更新日期:2020-04-13
中文翻译:

微生物菌落表型结构的基因工程控制。
细胞工程 1,2 的快速发展使合成生物学能够解决治疗 3,4 和工业 5 问题,但一个重大障碍是在异质的现实世界环境(如肠道 6,7、实体瘤 8、9、生物反应器 10)中出现无数意料之外的细胞反应或土壤 11。环境和细胞之间的复杂相互作用通常是通过不均匀的营养可用性产生的,当细胞通过表型分化12、13 适应和改变其局部环境时,这会产生双向耦合。尽管已在均质条件下探索了合成空间基因表达模式14-17,但基因回路、生长表型和环境的相互作用仍然是一个挑战。这里,我们设计的基因电路可以感知和控制包含生长和休眠细菌的微菌落中的表型结构。我们通过将不同的下游模块耦合到一个可调节的传感器来实现结构调制,该传感器利用大肠杆菌的应激反应并在生长停滞时被激活。一种是减慢生长并因此改变营养梯度的执行器模块。正如时空计算模型所预测的那样,该电路中的环境反馈会在菌落内部产生生长和休眠之间的稳健循环。我们还使用传感器驱动一个可诱导的门控模块,用于在非分裂细胞中进行选择性基因表达,这使我们能够通过使用“压力门控裂解电路”消除休眠表型来从根本上改变种群结构。











































 京公网安备 11010802027423号
京公网安备 11010802027423号