当前位置:
X-MOL 学术
›
Microsc. Res. Tech.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Metabolic labeling of Escherichia coli genomic DNA with erythrosine-11-dUTP for functional imaging via correlative microscopy.
Microscopy Research and Technique ( IF 2.0 ) Pub Date : 2020-03-31 , DOI: 10.1002/jemt.23487 Alexandre Loukanov 1, 2 , Svetla Nikolova 3 , Chavdar Filipov 4 , Seiichiro Nakabayashi 1
Microscopy Research and Technique ( IF 2.0 ) Pub Date : 2020-03-31 , DOI: 10.1002/jemt.23487 Alexandre Loukanov 1, 2 , Svetla Nikolova 3 , Chavdar Filipov 4 , Seiichiro Nakabayashi 1
Affiliation
|
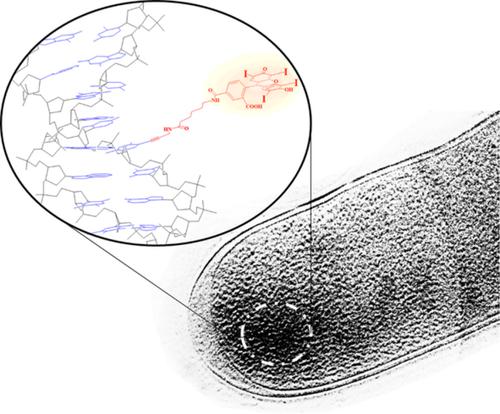
The fluorescent metabolic labeling of microorganisms genome is an advanced imaging technique to observe and study the native shapes, structural changes, functions, and tracking of nucleic acids in single cells or tissues. We have attempted to visualize the newly synthesized DNA within the intact nucleoid of ice‐embedded proliferating cells of Escherichia coli K‐12 (thymidine‐requiring mutant, strain N4316) via correlative light‐electron microscopy. For that purpose, erythrosine‐11‐dUTP was synthesized and used as a modified analog of the exogenous thymidine substrate for metabolic incorporation into the bacterial chromosome. The formed fluorescent genomic DNA during in cellulo polymerase reaction caused a minimal cellular arrest and cytotoxicity of E. coli at certain controlled conditions. The stained cells were visualized in typical red emission color via an epifluorescence microscope. They were further ice‐embedded and examined with a Hilbert differential contrast transmission electron microscopy. At high‐resolution, the ultrastructure of tagged nucleoid appeared with significantly higher electron dense in comparison to the unlabeled one. The enhanced contrast areas in the chromosome were ascribed to the presence of iodine contents from erythrosine dye. The presented labeling approach might be a powerful strategy to reveal the structural and dynamic changes in natural DNA replication including the relationship between newly synthesized in vivo nucleic acid and the physiological state of the cell.
中文翻译:

赤霉素11 dUTP对大肠杆菌基因组DNA的代谢标记,用于通过相关显微镜进行功能成像。
微生物基因组的荧光代谢标记是一种先进的成像技术,用于观察和研究单个细胞或组织中的天然形状,结构变化,功能和核酸追踪。我们试图通过相关的光电子显微镜观察大肠埃希氏菌K-12(需胸腺嘧啶核苷的突变体,菌株N4316)的冰包埋增殖细胞完整核苷酸中的新合成DNA 。为此目的,合成了赤藓氨酸-11-dUTP,并用作外源胸苷底物的修饰类似物,用于代谢结合到细菌染色体中。在纤维素聚合酶反应过程中形成的荧光基因组DNA导致最小的细胞停滞和大肠杆菌的细胞毒性在某些受控条件下。通过落射荧光显微镜将染色的细胞以典型的红色发射颜色可视化。将它们进一步冰包埋,并用希尔伯特差示透射电子显微镜检查。在高分辨率下,与未标记的相比,标记的核苷的超微结构具有更高的电子密度。染色体中对比度增强的区域归因于赤藓红染料中碘含量的存在。提出的标记方法可能是揭示天然DNA复制中结构和动态变化的强大策略,其中包括新合成的体内核酸与细胞生理状态之间的关系。
更新日期:2020-03-31
中文翻译:

赤霉素11 dUTP对大肠杆菌基因组DNA的代谢标记,用于通过相关显微镜进行功能成像。
微生物基因组的荧光代谢标记是一种先进的成像技术,用于观察和研究单个细胞或组织中的天然形状,结构变化,功能和核酸追踪。我们试图通过相关的光电子显微镜观察大肠埃希氏菌K-12(需胸腺嘧啶核苷的突变体,菌株N4316)的冰包埋增殖细胞完整核苷酸中的新合成DNA 。为此目的,合成了赤藓氨酸-11-dUTP,并用作外源胸苷底物的修饰类似物,用于代谢结合到细菌染色体中。在纤维素聚合酶反应过程中形成的荧光基因组DNA导致最小的细胞停滞和大肠杆菌的细胞毒性在某些受控条件下。通过落射荧光显微镜将染色的细胞以典型的红色发射颜色可视化。将它们进一步冰包埋,并用希尔伯特差示透射电子显微镜检查。在高分辨率下,与未标记的相比,标记的核苷的超微结构具有更高的电子密度。染色体中对比度增强的区域归因于赤藓红染料中碘含量的存在。提出的标记方法可能是揭示天然DNA复制中结构和动态变化的强大策略,其中包括新合成的体内核酸与细胞生理状态之间的关系。











































 京公网安备 11010802027423号
京公网安备 11010802027423号