Nature ( IF 50.5 ) Pub Date : 2020-04-08 , DOI: 10.1038/s41586-020-2188-x Katja Luck 1, 2, 3 , Dae-Kyum Kim 1, 4, 5, 6 , Luke Lambourne 1, 2, 3 , Kerstin Spirohn 1, 2, 3 , Bridget E Begg 1, 2, 3 , Wenting Bian 1, 2, 3 , Ruth Brignall 1, 2, 3 , Tiziana Cafarelli 1, 2, 3 , Francisco J Campos-Laborie 7, 8 , Benoit Charloteaux 1, 2, 3 , Dongsic Choi 9 , Atina G Coté 1, 4, 5, 6 , Meaghan Daley 1, 2, 3 , Steven Deimling 10 , Alice Desbuleux 1, 2, 3, 11 , Amélie Dricot 1, 2, 3 , Marinella Gebbia 1, 4, 5, 6 , Madeleine F Hardy 1, 2, 3 , Nishka Kishore 1, 4, 5, 6 , Jennifer J Knapp 1, 4, 5, 6 , István A Kovács 1, 12, 13 , Irma Lemmens 14, 15 , Miles W Mee 4, 5, 16 , Joseph C Mellor 1, 4, 5, 6, 17 , Carl Pollis 1, 2, 3 , Carles Pons 18 , Aaron D Richardson 1, 2, 3 , Sadie Schlabach 1, 2, 3 , Bridget Teeking 1, 2, 3 , Anupama Yadav 1, 2, 3 , Mariana Babor 1, 4, 5, 6 , Dawit Balcha 1, 2, 3 , Omer Basha 19, 20 , Christian Bowman-Colin 2, 3 , Suet-Feung Chin 21 , Soon Gang Choi 1, 2, 3 , Claudia Colabella 22, 23 , Georges Coppin 1, 2, 3, 11 , Cassandra D'Amata 10 , David De Ridder 1, 2, 3 , Steffi De Rouck 14, 15 , Miquel Duran-Frigola 18 , Hanane Ennajdaoui 1, 4, 5, 6 , Florian Goebels 4, 5, 16 , Liana Goehring 2, 3 , Anjali Gopal 1, 4, 5, 6 , Ghazal Haddad 1, 4, 5, 6 , Elodie Hatchi 2, 3 , Mohamed Helmy 4, 5, 16 , Yves Jacob 24, 25 , Yoseph Kassa 1, 2, 3 , Serena Landini 2, 3 , Roujia Li 1, 4, 5, 6 , Natascha van Lieshout 1, 4, 5, 6 , Andrew MacWilliams 1, 2, 3 , Dylan Markey 1, 2, 3 , Joseph N Paulson 26, 27, 28 , Sudharshan Rangarajan 1, 2, 3 , John Rasla 1, 2, 3 , Ashyad Rayhan 1, 4, 5, 6 , Thomas Rolland 1, 2, 3 , Adriana San-Miguel 1, 2, 3 , Yun Shen 1, 2, 3 , Dayag Sheykhkarimli 1, 4, 5, 6 , Gloria M Sheynkman 1, 2, 3 , Eyal Simonovsky 19, 20 , Murat Taşan 1, 4, 5, 6, 16 , Alexander Tejeda 1, 2, 3 , Vincent Tropepe 10 , Jean-Claude Twizere 11 , Yang Wang 1, 2, 3 , Robert J Weatheritt 4 , Jochen Weile 1, 4, 5, 6, 16 , Yu Xia 1, 29 , Xinping Yang 1, 2, 3 , Esti Yeger-Lotem 19, 20 , Quan Zhong 1, 2, 3, 30 , Patrick Aloy 18, 31 , Gary D Bader 4, 5, 16 , Javier De Las Rivas 7, 8 , Suzanne Gaudet 1, 2, 3 , Tong Hao 1, 2, 3 , Janusz Rak 9 , Jan Tavernier 14, 15 , David E Hill 1, 2, 3 , Marc Vidal 1, 2 , Frederick P Roth 1, 4, 5, 6, 16, 32 , Michael A Calderwood 1, 2, 3
|
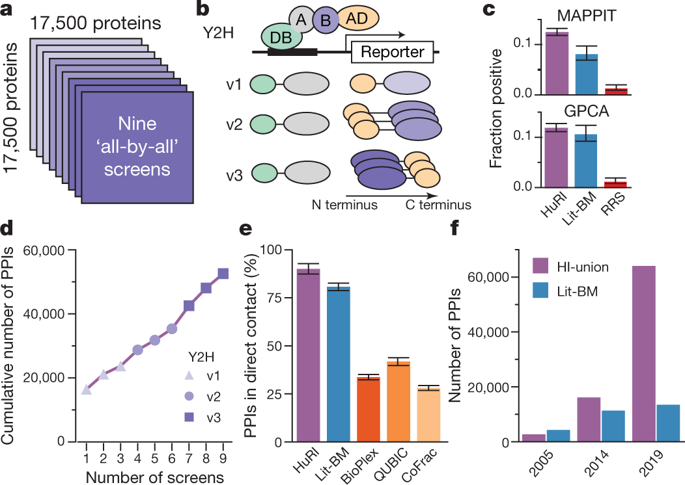
Global insights into cellular organization and genome function require comprehensive understanding of the interactome networks that mediate genotype–phenotype relationships1,2. Here we present a human ‘all-by-all’ reference interactome map of human binary protein interactions, or ‘HuRI’. With approximately 53,000 protein–protein interactions, HuRI has approximately four times as many such interactions as there are high-quality curated interactions from small-scale studies. The integration of HuRI with genome3, transcriptome4 and proteome5 data enables cellular function to be studied within most physiological or pathological cellular contexts. We demonstrate the utility of HuRI in identifying the specific subcellular roles of protein–protein interactions. Inferred tissue-specific networks reveal general principles for the formation of cellular context-specific functions and elucidate potential molecular mechanisms that might underlie tissue-specific phenotypes of Mendelian diseases. HuRI is a systematic proteome-wide reference that links genomic variation to phenotypic outcomes.
中文翻译:

人类二元蛋白相互作用组的参考图
对细胞组织和基因组功能的全球洞察需要全面了解介导基因型-表型关系1,2的相互作用组网络。在这里,我们提出了人类二元蛋白质相互作用或“HuRI”的人类“全部”参考相互作用组图。HuRI 有大约 53,000 种蛋白质-蛋白质相互作用,其此类相互作用的数量大约是小规模研究中高质量策划的相互作用的四倍。HuRI与基因组3、转录组4和蛋白质组5的整合数据使细胞功能能够在大多数生理或病理细胞环境中进行研究。我们证明了 HuRI 在识别蛋白质-蛋白质相互作用的特定亚细胞作用方面的效用。推断的组织特异性网络揭示了形成细胞环境特异性功能的一般原则,并阐明了可能构成孟德尔疾病组织特异性表型的潜在分子机制。HuRI 是一个系统的蛋白质组参考,将基因组变异与表型结果联系起来。











































 京公网安备 11010802027423号
京公网安备 11010802027423号