当前位置:
X-MOL 学术
›
J. Zool. Syst. Evol. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Telomere structure in insects: A review
Journal of Zoological Systematics and Evolutionary Research ( IF 2.0 ) Pub Date : 2019-11-19 , DOI: 10.1111/jzs.12332 Valentina Kuznetsova 1 , Snejana Grozeva 2 , Vladimir Gokhman 3
Journal of Zoological Systematics and Evolutionary Research ( IF 2.0 ) Pub Date : 2019-11-19 , DOI: 10.1111/jzs.12332 Valentina Kuznetsova 1 , Snejana Grozeva 2 , Vladimir Gokhman 3
Affiliation
|
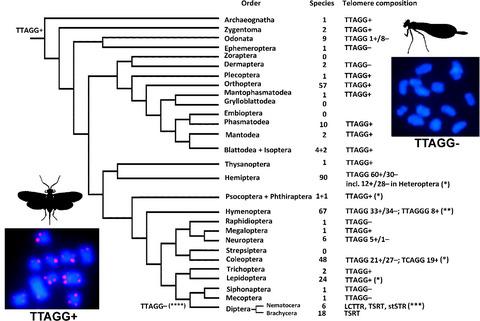
Telomeres are terminal regions of chromosomes, which protect them from fusion with other chromosomes and stabilize their structure. Telomeres usually contain specific DNA repeats (motifs), which are maintained by telomerase, a kind of reverse transcriptase. In this review, we survey the current state of knowledge of telomere motifs in insects. Among Hexapoda, data on telomere composition are available for more than 350 species from 108 families and 25 orders. The telomere motif (TTAGG)n is considered ancestral for the class Insecta. However, certain insects have different and often unknown telomeric sequences. This apparently happens because telomerase‐dependent mechanisms usually coexist with various means of alternative lengthening of telomeres as backup mechanisms of telomere maintenance. This coexistence can explain losses and reappearances of the TTAGG repeat and telomerase‐dependent telomere replication in insect evolution. For example, a few higher taxa, such as Heteroptera (Hemiptera) and Hymenoptera, show presence of the (TTAGG)n motif in their basal clades as well as a subsequent loss and, at least in the Hymenoptera, independent reappearance of this repeat in some advanced groups. Analogously, most members of Coleoptera also retain the TTAGG repeat, although it is changed to TCAGG in certain families. Furthermore, the (TTAGG)n motif seems to have been irreversibly lost in the order Diptera. In this group, telomeric sequences are represented either by long terminal repeats or by retrotransposons. Retrotransposons are also interspersed with other telomeric sequences in many groups of insects. The accumulating data demonstrate that the class Insecta is substantially diverse in terms of its telomere structure.
中文翻译:

昆虫端粒结构的研究进展
端粒是染色体的末端区域,可保护端粒不与其他染色体融合并稳定其结构。端粒通常包含特定的DNA重复序列(基序),这些重复序列由端粒酶(一种逆转录酶)维持。在这篇综述中,我们调查了昆虫端粒基序的当前知识状态。在六足纲中,有来自108个科和25个目的350多个物种的端粒组成数据。端粒基序(TTAGG)n被认为是Insecta类的祖先。但是,某些昆虫具有不同且通常未知的端粒序列。这显然是因为端粒酶依赖性机制通常与端粒替代性延长的各种手段共存,作为端粒维持的备用机制。这种共存可以解释昆虫进化过程中TTAGG重复序列和端粒酶依赖性端粒复制的损失和再现。例如,一些较高的分类单元,例如异翅目(Hemiptera)和膜翅目(Hymenoptera),显示出(TTAGG)n在其基部进化枝中的基序以及随后的损失,至少在膜翅目中,在某些先进群体中这种重复的独立再现。类似地,鞘翅目的大多数成员也保留了TTAGG重复序列,尽管在某些家族中已将其更改为TCAGG。此外,(TTAGG)n基序似乎已按双翅目顺序不可逆地丢失。在这一组中,端粒序列由长末端重复序列或反转录转座子代表。逆转座子还散布在许多昆虫群中的其他端粒序列中。不断积累的数据表明,昆虫纲昆虫的端粒结构实质上是多种多样的。
更新日期:2019-11-19
中文翻译:

昆虫端粒结构的研究进展
端粒是染色体的末端区域,可保护端粒不与其他染色体融合并稳定其结构。端粒通常包含特定的DNA重复序列(基序),这些重复序列由端粒酶(一种逆转录酶)维持。在这篇综述中,我们调查了昆虫端粒基序的当前知识状态。在六足纲中,有来自108个科和25个目的350多个物种的端粒组成数据。端粒基序(TTAGG)n被认为是Insecta类的祖先。但是,某些昆虫具有不同且通常未知的端粒序列。这显然是因为端粒酶依赖性机制通常与端粒替代性延长的各种手段共存,作为端粒维持的备用机制。这种共存可以解释昆虫进化过程中TTAGG重复序列和端粒酶依赖性端粒复制的损失和再现。例如,一些较高的分类单元,例如异翅目(Hemiptera)和膜翅目(Hymenoptera),显示出(TTAGG)n在其基部进化枝中的基序以及随后的损失,至少在膜翅目中,在某些先进群体中这种重复的独立再现。类似地,鞘翅目的大多数成员也保留了TTAGG重复序列,尽管在某些家族中已将其更改为TCAGG。此外,(TTAGG)n基序似乎已按双翅目顺序不可逆地丢失。在这一组中,端粒序列由长末端重复序列或反转录转座子代表。逆转座子还散布在许多昆虫群中的其他端粒序列中。不断积累的数据表明,昆虫纲昆虫的端粒结构实质上是多种多样的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号