iScience ( IF 4.6 ) Pub Date : 2020-04-05 , DOI: 10.1016/j.isci.2020.101034 Zhengyu Luo 1 , Xiaorong Wang 2 , Hong Jiang 1 , Ruoyu Wang 3 , Jian Chen 4 , Yusheng Chen 5 , Qianlan Xu 1 , Jun Cao 1 , Xiaowen Gong 6 , Ji Wu 6 , Yungui Yang 5 , Wenbo Li 7 , Chunsheng Han 4 , C Yan Cheng 8 , Michael G Rosenfeld 9 , Fei Sun 2 , Xiaoyuan Song 1
|
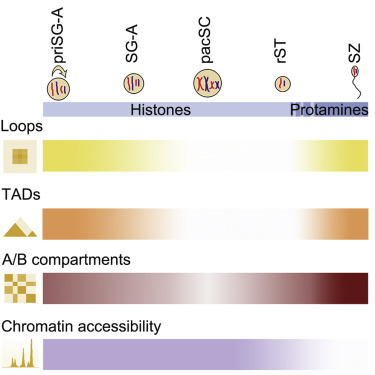
Three-dimensional chromatin structures undergo dynamic reorganization during mammalian spermatogenesis; however, their impacts on gene regulation remain unclear. Here, we focused on understanding the structure-function regulation of meiotic chromosomes by Hi-C and other omics techniques in mouse spermatogenesis across five stages. Beyond confirming recent reports regarding changes in compartmentalization and reorganization of topologically associating domains (TADs), we further demonstrated that chromatin loops are present prior to and after, but not at, the pachytene stage. By integrating Hi-C and RNA-seq data, we showed that the switching of A/B compartments between spermatogenic stages is tightly associated with meiosis-specific mRNAs and piRNAs expression. Moreover, our ATAC-seq data indicated that chromatin accessibility per se is not responsible for the TAD and loop diminishment at pachytene. Additionally, our ChIP-seq data demonstrated that CTCF and cohesin remain bound at TAD boundary regions throughout meiosis, suggesting that dynamic reorganization of TADs does not require CTCF and cohesin clearance.
中文翻译:

重组的 3D 基因组结构支持小鼠精子发生的转录调控。
三维染色质结构在哺乳动物精子发生过程中经历动态重组;然而,它们对基因调控的影响仍不清楚。在这里,我们重点了解 Hi-C 和其他组学技术对小鼠精子发生五个阶段减数分裂染色体的结构功能调控。除了证实最近有关拓扑关联域(TAD)的区室化和重组变化的报道之外,我们还进一步证明,染色质环存在于粗线期之前和之后,但不存在于粗线期。通过整合 Hi-C 和 RNA-seq 数据,我们发现生精阶段之间 A/B 区室的切换与减数分裂特异性 mRNA 和 piRNA 表达密切相关。此外,我们的 ATAC-seq 数据表明,染色质可及性本身并不导致粗线期的 TAD 和环减少。此外,我们的 ChIP-seq 数据表明,CTCF 和粘连蛋白在整个减数分裂过程中仍结合在 TAD 边界区域,这表明 TAD 的动态重组不需要 CTCF 和粘连蛋白清除。











































 京公网安备 11010802027423号
京公网安备 11010802027423号