当前位置:
X-MOL 学术
›
ACS Comb. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
In Vitro Identification of the Hamiltonian Cycle Using a Circular Structure Assisted DNA Computer.
ACS Combinatorial Science ( IF 3.903 ) Pub Date : 2020-04-06 , DOI: 10.1021/acscombsci.9b00150 Deepak Sharma 1 , Manojkumar Ramteke 1
ACS Combinatorial Science ( IF 3.903 ) Pub Date : 2020-04-06 , DOI: 10.1021/acscombsci.9b00150 Deepak Sharma 1 , Manojkumar Ramteke 1
Affiliation
|
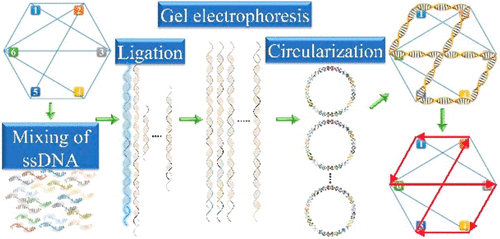
Adleman's illustration of molecular computing using DNA paved the way toward an entirely new direction of computing (Adleman, L. M. Science 1994, 266, 1021). The exponential time complex combinatorial problem on a traditional computer turns out to be a separation problem involving a polynomial number of steps in DNA computing experiments. Despite being a promising concept, the implementations of existing DNA computing procedures were restricted only to the smaller size formulations. In this work, we demonstrate a structure assisted DNA computing procedure on a bigger size Hamiltonian cycle problem involving 18 vertices. The developed model involves the formation and digestion of circular structure DNA, iteratively over multiple stages to eliminate the incorrect solutions to the given combinatorial problem. A high accuracy is obtained compared to other structure assisted models, which enable one to solve the bigger size problems.
中文翻译:

使用圆形结构辅助DNA计算机体外鉴定哈密顿循环。
阿德曼(Adleman)对使用DNA进行分子计算的插图为计算的全新方向铺平了道路(Adleman,LM Science 1994,266,1021)。传统计算机上的指数时间复杂组合问题被证明是一个分离问题,涉及DNA计算实验中多项式步数。尽管是一个有前途的概念,但现有DNA计算程序的实现仅限于较小尺寸的配方。在这项工作中,我们展示了涉及18个顶点的更大尺寸哈密顿循环问题的结构辅助DNA计算程序。所开发的模型涉及环状结构DNA的形成和消化,在多个阶段中反复进行,以消除针对给定组合问题的错误解决方案。
更新日期:2020-03-26
中文翻译:

使用圆形结构辅助DNA计算机体外鉴定哈密顿循环。
阿德曼(Adleman)对使用DNA进行分子计算的插图为计算的全新方向铺平了道路(Adleman,LM Science 1994,266,1021)。传统计算机上的指数时间复杂组合问题被证明是一个分离问题,涉及DNA计算实验中多项式步数。尽管是一个有前途的概念,但现有DNA计算程序的实现仅限于较小尺寸的配方。在这项工作中,我们展示了涉及18个顶点的更大尺寸哈密顿循环问题的结构辅助DNA计算程序。所开发的模型涉及环状结构DNA的形成和消化,在多个阶段中反复进行,以消除针对给定组合问题的错误解决方案。



























 京公网安备 11010802027423号
京公网安备 11010802027423号