Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A molecular simulation approach towards the development of universal nanocarriers by studying the pH- and electrostatic-driven changes in the dynamic structure of albumin
RSC Advances ( IF 3.9 ) Pub Date : 2020-4-2 , DOI: 10.1039/d0ra00803f Amit Kumar Srivastav 1 , Sanjeev K Gupta 2 , Umesh Kumar 1
RSC Advances ( IF 3.9 ) Pub Date : 2020-4-2 , DOI: 10.1039/d0ra00803f Amit Kumar Srivastav 1 , Sanjeev K Gupta 2 , Umesh Kumar 1
Affiliation
|
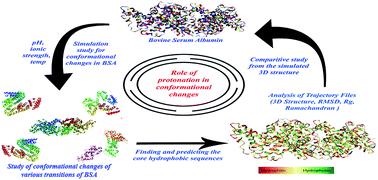
To explore the intramolecular interactions of protein, and its folding and unfolding mechanisms, we performed a simulation-based comparative study on albumin at different ionic strengths and pH. In this study, we performed molecular dynamics (MD) simulation for bovine serum albumin (BSA) at five different concentrations of NaCl (10, 20, 30, 40 and 50 mM), and five different pH values (2.0, 3.5, 4.3, 7.4, and 9.0). Herein, our aim was to unravel the effects of both pH and ionic strength on the conformations of the serum albumin structure. Our results indicate the effects of physicochemical factors in promoting conformational changes in the albumin structure, unlocking the hydrophobic sequences for hydrophobic drug binding. The BSA structure showed similarity to its native state in the pH range of 4.5 to 7.4 and at various ionic concentrations of NaCl. In the pH range of 3.5 to 4.5, the BSA structure showed denaturation in a controlled manner, which caused significant conformational changes in the molecular position of its hydrophobic amino acid residues. The resultant 3D structure gives insight into the amino acid trajectories. High denaturation and unstable behavior in the structural and conformational changes of the protein structure were observed at pH 2.0 and pH 9.0. We believe that these results and conditions will be helpful in the development of protein-based universal nanocarriers for the encapsulation of both hydrophilic and hydrophobic drugs.
中文翻译:

通过研究白蛋白动态结构的 pH 和静电驱动变化来开发通用纳米载体的分子模拟方法
为了探索蛋白质的分子内相互作用及其折叠和展开机制,我们对不同离子强度和 pH 值的白蛋白进行了基于模拟的比较研究。在本研究中,我们在五种不同浓度的 NaCl(10、20、30、40 和 50 mM)和五种不同的 pH 值(2.0、3.5、4.3、 7.4 和 9.0)。在此,我们的目标是揭示 pH 值和离子强度对血清白蛋白结构构象的影响。我们的结果表明物理化学因素在促进白蛋白结构构象变化、解锁疏水序列以进行疏水药物结合方面的作用。BSA 结构在 4.5 至 7 的 pH 值范围内显示出与其天然状态的相似性。4 和不同离子浓度的 NaCl。在 3.5 至 4.5 的 pH 范围内,BSA 结构以受控方式显示变性,这导致其疏水氨基酸残基的分子位置发生显着的构象变化。由此产生的 3D 结构可以深入了解氨基酸轨迹。在 pH 2.0 和 pH 9.0 下观察到蛋白质结构的结构和构象变化的高度变性和不稳定行为。我们相信这些结果和条件将有助于开发基于蛋白质的通用纳米载体,用于包封亲水性和疏水性药物。这导致其疏水氨基酸残基的分子位置发生显着的构象变化。由此产生的 3D 结构可以深入了解氨基酸轨迹。在 pH 2.0 和 pH 9.0 下观察到蛋白质结构的结构和构象变化的高度变性和不稳定行为。我们相信这些结果和条件将有助于开发基于蛋白质的通用纳米载体,用于包封亲水性和疏水性药物。这导致其疏水氨基酸残基的分子位置发生显着的构象变化。由此产生的 3D 结构可以深入了解氨基酸轨迹。在 pH 2.0 和 pH 9.0 下观察到蛋白质结构的结构和构象变化的高度变性和不稳定行为。我们相信这些结果和条件将有助于开发基于蛋白质的通用纳米载体,用于包封亲水性和疏水性药物。
更新日期:2020-04-02
中文翻译:

通过研究白蛋白动态结构的 pH 和静电驱动变化来开发通用纳米载体的分子模拟方法
为了探索蛋白质的分子内相互作用及其折叠和展开机制,我们对不同离子强度和 pH 值的白蛋白进行了基于模拟的比较研究。在本研究中,我们在五种不同浓度的 NaCl(10、20、30、40 和 50 mM)和五种不同的 pH 值(2.0、3.5、4.3、 7.4 和 9.0)。在此,我们的目标是揭示 pH 值和离子强度对血清白蛋白结构构象的影响。我们的结果表明物理化学因素在促进白蛋白结构构象变化、解锁疏水序列以进行疏水药物结合方面的作用。BSA 结构在 4.5 至 7 的 pH 值范围内显示出与其天然状态的相似性。4 和不同离子浓度的 NaCl。在 3.5 至 4.5 的 pH 范围内,BSA 结构以受控方式显示变性,这导致其疏水氨基酸残基的分子位置发生显着的构象变化。由此产生的 3D 结构可以深入了解氨基酸轨迹。在 pH 2.0 和 pH 9.0 下观察到蛋白质结构的结构和构象变化的高度变性和不稳定行为。我们相信这些结果和条件将有助于开发基于蛋白质的通用纳米载体,用于包封亲水性和疏水性药物。这导致其疏水氨基酸残基的分子位置发生显着的构象变化。由此产生的 3D 结构可以深入了解氨基酸轨迹。在 pH 2.0 和 pH 9.0 下观察到蛋白质结构的结构和构象变化的高度变性和不稳定行为。我们相信这些结果和条件将有助于开发基于蛋白质的通用纳米载体,用于包封亲水性和疏水性药物。这导致其疏水氨基酸残基的分子位置发生显着的构象变化。由此产生的 3D 结构可以深入了解氨基酸轨迹。在 pH 2.0 和 pH 9.0 下观察到蛋白质结构的结构和构象变化的高度变性和不稳定行为。我们相信这些结果和条件将有助于开发基于蛋白质的通用纳米载体,用于包封亲水性和疏水性药物。











































 京公网安备 11010802027423号
京公网安备 11010802027423号