Nature Communications ( IF 14.7 ) Pub Date : 2020-04-03 , DOI: 10.1038/s41467-020-15428-0 Nikolaus A Watson 1 , Tyrell N Cartwright 1 , Conor Lawless 2 , Marcos Cámara-Donoso 1 , Onur Sen 1 , Kosuke Sako 3 , Toru Hirota 3 , Hiroshi Kimura 4 , Jonathan M G Higgins 1
|
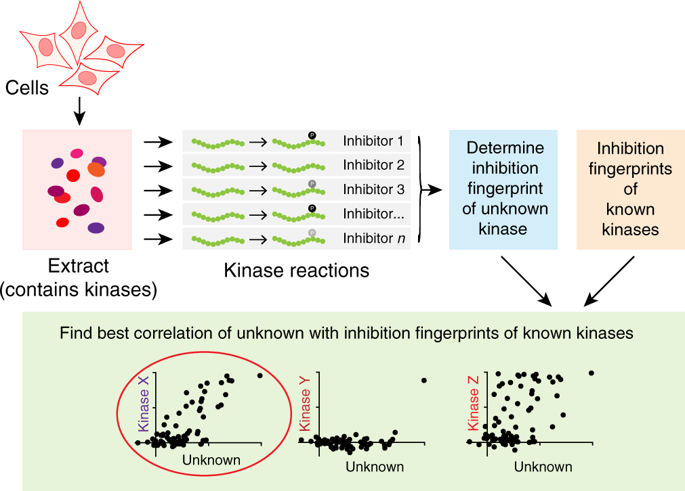
There are thousands of known cellular phosphorylation sites, but the paucity of ways to identify kinases for particular phosphorylation events remains a major roadblock for understanding kinase signaling. To address this, we here develop a generally applicable method that exploits the large number of kinase inhibitors that have been profiled on near-kinome-wide panels of protein kinases. The inhibition profile for each kinase provides a fingerprint that allows identification of unknown kinases acting on target phosphosites in cell extracts. We validate the method on diverse known kinase-phosphosite pairs, including histone kinases, EGFR autophosphorylation, and Integrin β1 phosphorylation by Src-family kinases. We also use our approach to identify the previously unknown kinases responsible for phosphorylation of INCENP at a site within a commonly phosphorylated motif in mitosis (a non-canonical target of Cyclin B-Cdk1), and of BCL9L at S915 (PKA). We show that the method has clear advantages over in silico and genetic screening.
中文翻译:

激酶抑制谱作为识别特定磷酸化位点激酶的工具。
已知的细胞磷酸化位点有数千个,但缺乏识别特定磷酸化事件激酶的方法仍然是理解激酶信号传导的主要障碍。为了解决这个问题,我们在这里开发了一种普遍适用的方法,该方法利用已在近激酶组范围的蛋白激酶组上进行分析的大量激酶抑制剂。每种激酶的抑制谱提供了指纹,可以识别作用于细胞提取物中的靶磷酸位点的未知激酶。我们在多种已知的激酶-磷酸位点对上验证了该方法,包括组蛋白激酶、EGFR 自磷酸化和 Src 家族激酶的整合素 β1 磷酸化。我们还使用我们的方法来识别以前未知的激酶,这些激酶负责有丝分裂中常见磷酸化基序(细胞周期蛋白 B-Cdk1 的非规范靶点)内的 INCENP 磷酸化以及 S915 (PKA) 上的 BCL9L 磷酸化。我们证明该方法比计算机和基因筛选具有明显的优势。











































 京公网安备 11010802027423号
京公网安备 11010802027423号