当前位置:
X-MOL 学术
›
Nat. Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer.
Nature Genetics ( IF 31.7 ) Pub Date : 2020-04-03 , DOI: 10.1038/s41588-020-0603-8 Alex H Wagner 1 , Brian Walsh 2 , Georgia Mayfield 2 , David Tamborero 3, 4 , Dmitriy Sonkin 5 , Kilannin Krysiak 1 , Jordi Deu-Pons 6, 7 , Ryan P Duren 8 , Jianjiong Gao 9 , Julie McMurry 2 , Sara Patterson 10 , Catherine Del Vecchio Fitz 11 , Beth A Pitel 12 , Ozman U Sezerman 13 , Kyle Ellrott 2 , Jeremy L Warner 14 , Damian T Rieke 15 , Tero Aittokallio 16, 17 , Ethan Cerami 11 , Deborah I Ritter 18, 19 , Lynn M Schriml 20 , Robert R Freimuth 12 , Melissa Haendel 2, 21 , Gordana Raca 22, 23 , Subha Madhavan 24 , Michael Baudis 25 , Jacques S Beckmann 26 , Rodrigo Dienstmann 27 , Debyani Chakravarty 9 , Xuan Shirley Li 8 , Susan Mockus 10 , Olivier Elemento 28 , Nikolaus Schultz 9 , Nuria Lopez-Bigas 3, 6, 7 , Mark Lawler 29 , Jeremy Goecks 2 , Malachi Griffith 1 , Obi L Griffith 1 , Adam A Margolin 2 ,
Nature Genetics ( IF 31.7 ) Pub Date : 2020-04-03 , DOI: 10.1038/s41588-020-0603-8 Alex H Wagner 1 , Brian Walsh 2 , Georgia Mayfield 2 , David Tamborero 3, 4 , Dmitriy Sonkin 5 , Kilannin Krysiak 1 , Jordi Deu-Pons 6, 7 , Ryan P Duren 8 , Jianjiong Gao 9 , Julie McMurry 2 , Sara Patterson 10 , Catherine Del Vecchio Fitz 11 , Beth A Pitel 12 , Ozman U Sezerman 13 , Kyle Ellrott 2 , Jeremy L Warner 14 , Damian T Rieke 15 , Tero Aittokallio 16, 17 , Ethan Cerami 11 , Deborah I Ritter 18, 19 , Lynn M Schriml 20 , Robert R Freimuth 12 , Melissa Haendel 2, 21 , Gordana Raca 22, 23 , Subha Madhavan 24 , Michael Baudis 25 , Jacques S Beckmann 26 , Rodrigo Dienstmann 27 , Debyani Chakravarty 9 , Xuan Shirley Li 8 , Susan Mockus 10 , Olivier Elemento 28 , Nikolaus Schultz 9 , Nuria Lopez-Bigas 3, 6, 7 , Mark Lawler 29 , Jeremy Goecks 2 , Malachi Griffith 1 , Obi L Griffith 1 , Adam A Margolin 2 ,
Affiliation
|
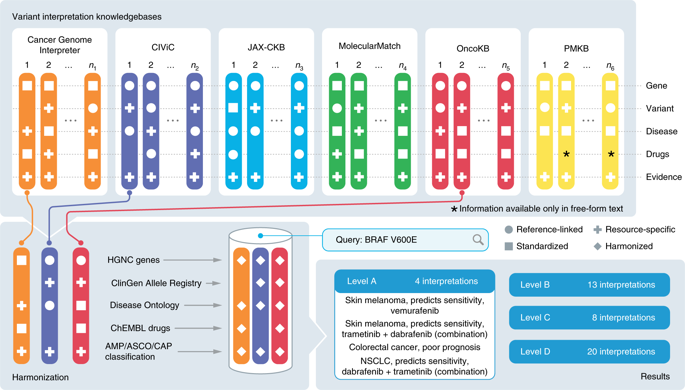
Precision oncology relies on accurate discovery and interpretation of genomic variants, enabling individualized diagnosis, prognosis and therapy selection. We found that six prominent somatic cancer variant knowledgebases were highly disparate in content, structure and supporting primary literature, impeding consensus when evaluating variants and their relevance in a clinical setting. We developed a framework for harmonizing variant interpretations to produce a meta-knowledgebase of 12,856 aggregate interpretations. We demonstrated large gains in overlap between resources across variants, diseases and drugs as a result of this harmonization. We subsequently demonstrated improved matching between a patient cohort and harmonized interpretations of potential clinical significance, observing an increase from an average of 33% per individual knowledgebase to 57% in aggregate. Our analyses illuminate the need for open, interoperable sharing of variant interpretation data. We also provide a freely available web interface (search.cancervariants.org) for exploring the harmonized interpretations from these six knowledgebases.
中文翻译:

癌症体细胞基因组变异临床解释的统一元知识库。
精准肿瘤学依赖于基因组变异的准确发现和解释,从而实现个体化诊断、预后和治疗选择。我们发现六个著名的体细胞癌症变异知识库在内容、结构和支持主要文献方面存在很大差异,这阻碍了在评估变异及其在临床环境中的相关性时达成共识。我们开发了一个协调不同解释的框架,以生成包含 12,856 种聚合解释的元知识库。由于这种协调,我们证明了跨变体、疾病和药物的资源之间的重叠取得了巨大的成果。随后,我们证明了患者队列与潜在临床意义的统一解释之间的匹配得到了改善,观察到每个知识库的平均 33% 增加到了总体 57%。我们的分析阐明了开放、可互操作共享变异解释数据的必要性。我们还提供免费的网络界面 (search.cancervariants.org),用于探索这六个知识库的统一解释。
更新日期:2020-04-24
中文翻译:

癌症体细胞基因组变异临床解释的统一元知识库。
精准肿瘤学依赖于基因组变异的准确发现和解释,从而实现个体化诊断、预后和治疗选择。我们发现六个著名的体细胞癌症变异知识库在内容、结构和支持主要文献方面存在很大差异,这阻碍了在评估变异及其在临床环境中的相关性时达成共识。我们开发了一个协调不同解释的框架,以生成包含 12,856 种聚合解释的元知识库。由于这种协调,我们证明了跨变体、疾病和药物的资源之间的重叠取得了巨大的成果。随后,我们证明了患者队列与潜在临床意义的统一解释之间的匹配得到了改善,观察到每个知识库的平均 33% 增加到了总体 57%。我们的分析阐明了开放、可互操作共享变异解释数据的必要性。我们还提供免费的网络界面 (search.cancervariants.org),用于探索这六个知识库的统一解释。











































 京公网安备 11010802027423号
京公网安备 11010802027423号