当前位置:
X-MOL 学术
›
Beilstein. J. Org. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Opening up connectivity between documents, structures and bioactivity
Beilstein Journal of Organic Chemistry ( IF 2.2 ) Pub Date : 2020-04-02 , DOI: 10.3762/bjoc.16.54 Christopher Southan 1, 2
Beilstein Journal of Organic Chemistry ( IF 2.2 ) Pub Date : 2020-04-02 , DOI: 10.3762/bjoc.16.54 Christopher Southan 1, 2
Affiliation
|
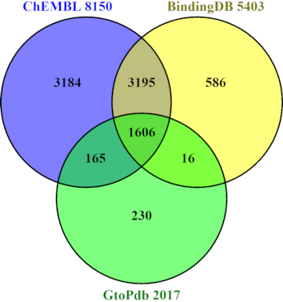
Bioscientists reading papers or patents strive to discern the key relationships reported within a document “D“ where a bioactivity “A” with a quantitative result “R” (e.g., an IC50) is reported for chemical structure “C” that modulates (e.g., inhibits) a protein target “P”. A useful shorthand for this connectivity thus becomes DARCP. The problem at the core of this article is that the community has spent millions effectively burying these relationships in PDFs over many decades but must now spend millions more trying to get them back out. The key imperative for this is to increase the flow into structured open databases. The positive impacts will include expanded data mining opportunities for drug discovery and chemical biology. Over the last decade commercial sources have manually extracted DARCP from ≈300,000 documents encompassing ≈7 million compounds interacting with ≈10,000 targets. Over a similar time, the Guide to Pharmacology, BindingDB and ChEMBL have carried out analogues DARCP extractions. Although their expert-curated numbers are lower (i.e., ≈2 million compounds against ≈3700 human proteins), these open sources have the great advantage of being merged within PubChem. Parallel efforts have focused on the extraction of document-to-compound (D-C-only) connectivity. In the absence of molecular mechanism of action (mmoa) annotation, this is of less value but can be automatically extracted. This has been significantly accomplished for patents, (e.g., by IBM, SureChEMBL and WIPO) for over 30 million compounds in PubChem. These have recently been joined by 1.4 million D-C submissions from three major chemistry publishers. In addition, both the European and US PubMed Central portals now add chemistry look-ups from abstracts and full-text papers. However, the fully automated extraction of DARCLP has not yet been achieved. This stands in contrast to the ability of biocurators to discern these relationships in minutes. Unfortunately, no journals have yet instigated a flow of author-specified DARCP directly into open databases. Progress may come from trends such as open science, open access (OA), findable, accessible, interoperable and reusable (FAIR), resource description framework (RDF) and WikiData. However, we will need to await the technical applicability in respect to DARCP capture to see if this opens up connectivity.
中文翻译:

打开文档、结构和生物活性之间的联系
阅读论文或专利的生物科学家努力辨别文件“D”中报告的关键关系,其中报告了具有定量结果“R”(例如 IC 50 )的生物活性“A”,用于调节化学结构“C”(例如,抑制)蛋白质靶标“P”。因此,这种连接的一个有用的简写方式就是 DARCP。本文的核心问题是,几十年来,社区花费了数百万美元有效地将这些关系隐藏在 PDF 中,但现在必须花费数百万美元试图将它们恢复出来。实现这一目标的关键是增加结构化开放数据库的流量。积极影响将包括扩大药物发现和化学生物学的数据挖掘机会。在过去的十年中,商业来源从约 300,000 个文档中手动提取了 DARCP,其中包含约 700 万个与约 10,000 个目标相互作用的化合物。与此同时,Guide to Pharmacology、BindingDB 和 ChEMBL 也进行了类似物 DARCP 提取。尽管专家策划的数量较低(即约 200 万种化合物对应约 3700 种人类蛋白质),但这些开源资源具有合并到 PubChem 中的巨大优势。并行工作的重点是提取文档到复合(仅限 DC)的连接。在缺乏分子作用机制(mmoa)注释的情况下,其价值较小,但可以自动提取。 PubChem 中超过 3000 万种化合物的专利(例如 IBM、SureChEMBL 和 WIPO)已取得显着成就。最近,来自三大化学出版商的 140 万份 DC 提交也加入其中。此外,欧洲和美国的 PubMed Central 门户现在都添加了摘要和全文论文的化学查找。 然而,DARCLP的全自动提取尚未实现。这与生物策展人在几分钟内辨别这些关系的能力形成鲜明对比。不幸的是,目前还没有期刊将作者指定的 DARCP 直接流入开放数据库。进步可能来自开放科学、开放获取(OA)、可查找、可访问、可互操作和可重用(FAIR)、资源描述框架(RDF)和维基数据等趋势。然而,我们需要等待 DARCP 捕获的技术适用性,看看这是否会打开连接。
更新日期:2020-04-03
中文翻译:

打开文档、结构和生物活性之间的联系
阅读论文或专利的生物科学家努力辨别文件“D”中报告的关键关系,其中报告了具有定量结果“R”(例如 IC 50 )的生物活性“A”,用于调节化学结构“C”(例如,抑制)蛋白质靶标“P”。因此,这种连接的一个有用的简写方式就是 DARCP。本文的核心问题是,几十年来,社区花费了数百万美元有效地将这些关系隐藏在 PDF 中,但现在必须花费数百万美元试图将它们恢复出来。实现这一目标的关键是增加结构化开放数据库的流量。积极影响将包括扩大药物发现和化学生物学的数据挖掘机会。在过去的十年中,商业来源从约 300,000 个文档中手动提取了 DARCP,其中包含约 700 万个与约 10,000 个目标相互作用的化合物。与此同时,Guide to Pharmacology、BindingDB 和 ChEMBL 也进行了类似物 DARCP 提取。尽管专家策划的数量较低(即约 200 万种化合物对应约 3700 种人类蛋白质),但这些开源资源具有合并到 PubChem 中的巨大优势。并行工作的重点是提取文档到复合(仅限 DC)的连接。在缺乏分子作用机制(mmoa)注释的情况下,其价值较小,但可以自动提取。 PubChem 中超过 3000 万种化合物的专利(例如 IBM、SureChEMBL 和 WIPO)已取得显着成就。最近,来自三大化学出版商的 140 万份 DC 提交也加入其中。此外,欧洲和美国的 PubMed Central 门户现在都添加了摘要和全文论文的化学查找。 然而,DARCLP的全自动提取尚未实现。这与生物策展人在几分钟内辨别这些关系的能力形成鲜明对比。不幸的是,目前还没有期刊将作者指定的 DARCP 直接流入开放数据库。进步可能来自开放科学、开放获取(OA)、可查找、可访问、可互操作和可重用(FAIR)、资源描述框架(RDF)和维基数据等趋势。然而,我们需要等待 DARCP 捕获的技术适用性,看看这是否会打开连接。











































 京公网安备 11010802027423号
京公网安备 11010802027423号