Methods ( IF 4.2 ) Pub Date : 2020-04-02 , DOI: 10.1016/j.ymeth.2020.03.008 Joshua D Karslake 1 , Eric D Donarski 1 , Sarah A Shelby 1 , Lucas M Demey 2 , Victor J DiRita 2 , Sarah L Veatch 1 , Julie S Biteen 3
|
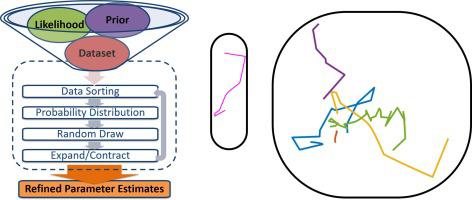
Single-molecule fluorescence microscopy probes nanoscale, subcellular biology in real time. Existing methods for analyzing single-particle tracking data provide dynamical information, but can suffer from supervisory biases and high uncertainties. Here, we develop a method for the case of multiple interconverting species undergoing free diffusion and introduce a new approach to analyzing single-molecule trajectories: the Single-Molecule Analysis by Unsupervised Gibbs sampling (SMAUG) algorithm, which uses nonparametric Bayesian statistics to uncover the whole range of information contained within a single-particle trajectory dataset. Even in complex systems where multiple biological states lead to a number of observed mobility states, SMAUG provides the number of mobility states, the average diffusion coefficient of single molecules in that state, the fraction of single molecules in that state, the localization noise, and the probability of transitioning between two different states. In this paper, we provide the theoretical background for the SMAUG analysis and then we validate the method using realistic simulations of single-particle trajectory datasets as well as experiments on a controlled in vitro system. Finally, we demonstrate SMAUG on real experimental systems in both prokaryotes and eukaryotes to measure the motions of the regulatory protein TcpP in Vibrio cholerae and the dynamics of the B-cell receptor antigen response pathway in lymphocytes. Overall, SMAUG provides a mathematically rigorous approach to measuring the real-time dynamics of molecular interactions in living cells.
中文翻译:

SMAUG:使用非参数贝叶斯统计分析单分子轨迹
单分子荧光显微镜实时探测纳米级亚细胞生物学。用于分析单粒子跟踪数据的现有方法提供动态信息,但可能会受到监督偏差和高不确定性的影响。在这里,我们针对多个相互转化的物种进行自由扩散的情况开发了一种方法,并介绍了一种分析单分子轨迹的新方法:通过无监督吉布斯采样 (SMAUG) 算法进行的单分子分析,该算法使用非参数贝叶斯统计来揭示包含在单粒子轨迹数据集中的全部信息。即使在多个生物状态导致许多观察到的迁移状态的复杂系统中,SMAUG 也提供迁移状态的数量,该状态下单个分子的平均扩散系数,处于该状态的单个分子的比例、定位噪声以及在两种不同状态之间转换的概率。在本文中,我们为 SMAUG 分析提供了理论背景,然后我们使用单粒子轨迹数据集的真实模拟以及受控实验验证了该方法。体外系统。最后,我们在原核生物和真核生物的真实实验系统上展示了 SMAUG,以测量霍乱弧菌中调节蛋白 TcpP 的运动和淋巴细胞中 B 细胞受体抗原反应途径的动力学。总体而言,SMAUG 提供了一种数学上严谨的方法来测量活细胞中分子相互作用的实时动态。











































 京公网安备 11010802027423号
京公网安备 11010802027423号