当前位置:
X-MOL 学术
›
Phys. Rev. X
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Toward a High-Resolution Reconstruction of 3D Nerve Fiber Architectures and Crossings in the Brain Using Light Scattering Measurements and Finite-Difference Time-Domain Simulations
Physical Review X ( IF 11.6 ) Pub Date : 2020-04-02 , DOI: 10.1103/physrevx.10.021002 Miriam Menzel , Markus Axer , Hans De Raedt , Irene Costantini , Ludovico Silvestri , Francesco S. Pavone , Katrin Amunts , Kristel Michielsen
Physical Review X ( IF 11.6 ) Pub Date : 2020-04-02 , DOI: 10.1103/physrevx.10.021002 Miriam Menzel , Markus Axer , Hans De Raedt , Irene Costantini , Ludovico Silvestri , Francesco S. Pavone , Katrin Amunts , Kristel Michielsen
|
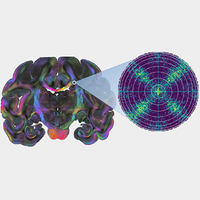
Unraveling the structure and function of the brain requires a detailed knowledge about the neuronal connections, i.e., the spatial architecture of the nerve fibers. One of the most powerful histological methods to reconstruct the three-dimensional nerve fiber pathways is 3D-polarized light imaging (3D-PLI). The technique measures the birefringence of histological brain sections and derives the spatial fiber orientations of whole human brain sections with micrometer resolution. However, the technique yields only a single fiber orientation for each measured tissue voxel even if it is composed of fibers with different orientations, so that in-plane crossing fibers are misinterpreted as out-of-plane fibers. When generating a detailed model of the three-dimensional nerve fiber architecture in the brain, a correct detection and interpretation of nerve fiber crossings is crucial. Here, we show how light scattering in transmission microscopy measurements can be leveraged to identify nerve fiber crossings in 3D-PLI data and demonstrate that measurements of the scattering pattern can resolve the substructure of brain tissue like the crossing angles of the nerve fibers. For this purpose, we develop a simulation framework that permits the study of transmission microscopy measurements—in particular, light scattering—on large-scale complex fiber structures like brain tissue, using finite-difference time-domain (FDTD) simulations and high-performance computing. The simulations are used not only to model and explain experimental observations, but also to develop new analysis methods and measurement techniques. We demonstrate in various experimental studies on brain sections from different species (rodent, monkey, and human) and in FDTD simulations that the polarization-independent transmitted light intensity (transmittance) decreases significantly (by more than 50%) with an increasing out-of-plane angle of the nerve fibers and that it is mostly independent of the in-plane crossing angle. Hence, the transmittance can be used to distinguish regions with low fiber density and in-plane crossing fibers from regions with out-of-plane fibers, solving a major problem in 3D-PLI and allowing for a much better reconstruction of the complex nerve fiber architecture in the brain. Finally, we show that light scattering (oblique illumination) in the visible spectrum reveals the underlying structure of brain tissue like the crossing angle of the nerve fibers with micrometer resolution, enabling an even more detailed reconstruction of nerve fiber crossings in the brain and opening up new fields of research.
中文翻译:

使用光散射测量和时域有限差分模拟,实现高分辨率的3D神经纤维结构和大脑中交叉的高分辨率重构。
弄清大脑的结构和功能需要有关神经元连接的详细知识,即神经纤维的空间结构。重建三维神经纤维通路的最强大的组织学方法之一是3D偏振光成像(3D-PLI)。该技术可测量组织学大脑切片的双折射,并以微米分辨率得出整个人类大脑切片的空间纤维方向。但是,该技术即使对每个测量的组织体素由具有不同方向的纤维组成,也只能产生单一的纤维方向,因此面内交叉纤维会被误解为面外纤维。当生成大脑中三维神经纤维结构的详细模型时,正确检测和解释神经纤维交叉至关重要。在这里,我们展示了如何利用透射显微镜测量中的光散射来识别3D-PLI数据中的神经纤维交叉,并证明散射模式的测量可以解决脑组织的子结构,如神经纤维的交叉角。为此,我们开发了一个模拟框架,该框架可以使用有限时域(FDTD)模拟和高性能研究在大型复杂纤维结构(例如脑组织)上进行透射显微镜测量,尤其是光散射的研究。计算。仿真不仅用于建模和解释实验观察结果,还用于开发新的分析方法和测量技术。我们在来自不同物种(啮齿动物,猴子和人类)的大脑切片的各种实验研究中以及在FDTD模拟中证明,与偏振无关的透射光强度(透射率)随着出射光的增加而显着降低(超过50%)。神经纤维的平面角,它主要与平面内交叉角无关。因此,透射率可用于区分纤维密度低和面内交叉纤维的区域与面外纤维的区域,从而解决了3D-PLI中的一个主要问题,并且可以更好地重建复杂的神经纤维大脑中的架构。最后,
更新日期:2020-04-02
中文翻译:

使用光散射测量和时域有限差分模拟,实现高分辨率的3D神经纤维结构和大脑中交叉的高分辨率重构。
弄清大脑的结构和功能需要有关神经元连接的详细知识,即神经纤维的空间结构。重建三维神经纤维通路的最强大的组织学方法之一是3D偏振光成像(3D-PLI)。该技术可测量组织学大脑切片的双折射,并以微米分辨率得出整个人类大脑切片的空间纤维方向。但是,该技术即使对每个测量的组织体素由具有不同方向的纤维组成,也只能产生单一的纤维方向,因此面内交叉纤维会被误解为面外纤维。当生成大脑中三维神经纤维结构的详细模型时,正确检测和解释神经纤维交叉至关重要。在这里,我们展示了如何利用透射显微镜测量中的光散射来识别3D-PLI数据中的神经纤维交叉,并证明散射模式的测量可以解决脑组织的子结构,如神经纤维的交叉角。为此,我们开发了一个模拟框架,该框架可以使用有限时域(FDTD)模拟和高性能研究在大型复杂纤维结构(例如脑组织)上进行透射显微镜测量,尤其是光散射的研究。计算。仿真不仅用于建模和解释实验观察结果,还用于开发新的分析方法和测量技术。我们在来自不同物种(啮齿动物,猴子和人类)的大脑切片的各种实验研究中以及在FDTD模拟中证明,与偏振无关的透射光强度(透射率)随着出射光的增加而显着降低(超过50%)。神经纤维的平面角,它主要与平面内交叉角无关。因此,透射率可用于区分纤维密度低和面内交叉纤维的区域与面外纤维的区域,从而解决了3D-PLI中的一个主要问题,并且可以更好地重建复杂的神经纤维大脑中的架构。最后,









































 京公网安备 11010802027423号
京公网安备 11010802027423号