Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Metagenomic views of microbial dynamics influenced by hydrocarbon seepage in sediments of the Gulf of Mexico.
Scientific Reports ( IF 3.8 ) Pub Date : 2020-04-01 , DOI: 10.1038/s41598-020-62840-z Rui Zhao 1 , Zarath M Summers 2 , Glenn D Christman 1 , Kristin M Yoshimura 1 , Jennifer F Biddle 1
Scientific Reports ( IF 3.8 ) Pub Date : 2020-04-01 , DOI: 10.1038/s41598-020-62840-z Rui Zhao 1 , Zarath M Summers 2 , Glenn D Christman 1 , Kristin M Yoshimura 1 , Jennifer F Biddle 1
Affiliation
|
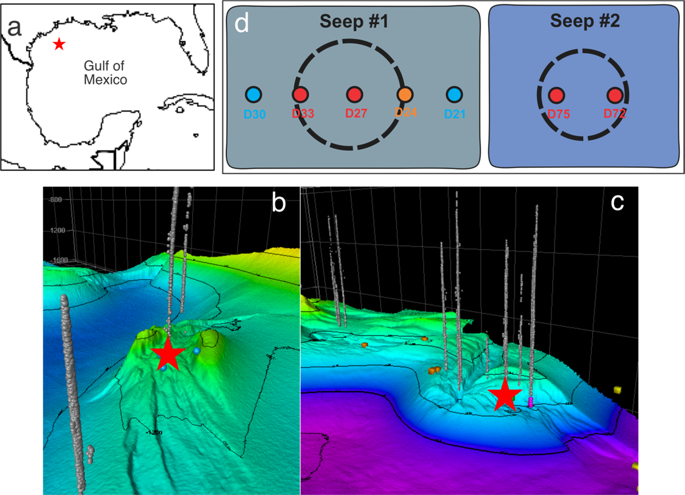
Microbial cells in the seabed are thought to persist by slow population turnover rates and extremely low energy requirements. External stimulations such as seafloor hydrocarbon seeps have been demonstrated to significantly boost microbial growth; however, the microbial community response has not been fully understood. Here we report a comparative metagenomic study of microbial response to natural hydrocarbon seeps in the Gulf of Mexico. Subsurface sediments (10-15 cm below seafloor) were collected from five natural seep sites and two reference sites. The resulting metagenome sequencing datasets were analyzed with both gene-based and genome-based approaches. 16S rRNA gene-based analyses suggest that the seep samples are distinct from the references by both 16S rRNA fractional content and phylogeny, with the former dominated by ANME-1 archaea (~50% of total) and Desulfobacterales, and the latter dominated by the Deltaproteobacteria, Planctomycetes, and Chloroflexi phyla. Sulfate-reducing bacteria (SRB) are present in both types of samples, with higher relative abundances in seep samples than the references. Genes for nitrogen fixation were predominantly found in the seep sites, whereas the reference sites showed a dominant signal for anaerobic ammonium oxidation (anammox). We recovered 49 metagenome-assembled genomes and assessed the microbial functional potentials in both types of samples. By this genome-based analysis, the seep samples were dominated by ANME-1 archaea and SRB, with the capacity for methane oxidation coupled to sulfate reduction, which is consistent with the 16S rRNA-gene based characterization. Although ANME-1 archaea and SRB are present in low relative abundances, genome bins from the reference sites are dominated by uncultured members of NC10 and anammox Scalindua, suggesting a prevalence of nitrogen transformations for energy in non-seep pelagic sediments. This study suggests that hydrocarbon seeps can greatly change the microbial community structure by stimulating nitrogen fixation, inherently shifting the nitrogen metabolism compared to those of the reference sediments.
中文翻译:

墨西哥湾沉积物中碳氢化合物渗漏影响的微生物动态宏基因组学观点。
人们认为海底的微生物细胞能够通过缓慢的种群周转率和极低的能量需求而持续存在。海底碳氢化合物渗漏等外部刺激已被证明可显着促进微生物生长;然而,微生物群落的反应尚未得到充分了解。在这里,我们报告了墨西哥湾微生物对天然碳氢化合物渗漏反应的比较宏基因组研究。从五个天然渗漏点和两个参考点收集地下沉积物(海底以下 10-15 厘米)。使用基于基因和基于基因组的方法对所得的宏基因组测序数据集进行了分析。基于 16S rRNA 基因的分析表明,渗液样品在 16S rRNA 分数含量和系统发育方面都与参考样品不同,前者以 ANME-1 古菌(约占总数的 50%)和脱硫杆菌为主,后者以Deltaproteobacteria、Planctomycetes 和 Chloroflexi 门。硫酸盐还原菌 (SRB) 存在于两种类型的样品中,渗液样品中的相对丰度高于参考样品。固氮基因主要发现于渗漏位点,而参考位点则显示出厌氧氨氧化(anammox)的主导信号。我们恢复了 49 个宏基因组组装的基因组,并评估了两种类型样本的微生物功能潜力。通过这种基于基因组的分析,渗漏样品以 ANME-1 古菌和 SRB 为主,具有甲烷氧化和硫酸盐还原能力,这与基于 16S rRNA 基因的表征一致。 尽管 ANME-1 古细菌和 SRB 的相对丰度较低,但参考位点的基因组箱主要由 NC10 和厌氧氨氧化斯卡林杜亚 (Anammox Scalindua) 的未培养成员主导,这表明非渗流中上层沉积物中普遍存在氮转化为能量的现象。这项研究表明,与参考沉积物相比,碳氢化合物渗漏可以通过刺激固氮来极大地改变微生物群落结构,从而从本质上改变氮代谢。
更新日期:2020-04-01
中文翻译:

墨西哥湾沉积物中碳氢化合物渗漏影响的微生物动态宏基因组学观点。
人们认为海底的微生物细胞能够通过缓慢的种群周转率和极低的能量需求而持续存在。海底碳氢化合物渗漏等外部刺激已被证明可显着促进微生物生长;然而,微生物群落的反应尚未得到充分了解。在这里,我们报告了墨西哥湾微生物对天然碳氢化合物渗漏反应的比较宏基因组研究。从五个天然渗漏点和两个参考点收集地下沉积物(海底以下 10-15 厘米)。使用基于基因和基于基因组的方法对所得的宏基因组测序数据集进行了分析。基于 16S rRNA 基因的分析表明,渗液样品在 16S rRNA 分数含量和系统发育方面都与参考样品不同,前者以 ANME-1 古菌(约占总数的 50%)和脱硫杆菌为主,后者以Deltaproteobacteria、Planctomycetes 和 Chloroflexi 门。硫酸盐还原菌 (SRB) 存在于两种类型的样品中,渗液样品中的相对丰度高于参考样品。固氮基因主要发现于渗漏位点,而参考位点则显示出厌氧氨氧化(anammox)的主导信号。我们恢复了 49 个宏基因组组装的基因组,并评估了两种类型样本的微生物功能潜力。通过这种基于基因组的分析,渗漏样品以 ANME-1 古菌和 SRB 为主,具有甲烷氧化和硫酸盐还原能力,这与基于 16S rRNA 基因的表征一致。 尽管 ANME-1 古细菌和 SRB 的相对丰度较低,但参考位点的基因组箱主要由 NC10 和厌氧氨氧化斯卡林杜亚 (Anammox Scalindua) 的未培养成员主导,这表明非渗流中上层沉积物中普遍存在氮转化为能量的现象。这项研究表明,与参考沉积物相比,碳氢化合物渗漏可以通过刺激固氮来极大地改变微生物群落结构,从而从本质上改变氮代谢。











































 京公网安备 11010802027423号
京公网安备 11010802027423号