当前位置:
X-MOL 学术
›
Adv. Synth. Catal.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Metagenomic Mining for Amine Dehydrogenase Discovery
Advanced Synthesis & Catalysis ( IF 5.4 ) Pub Date : 2020-03-30 , DOI: 10.1002/adsc.202000094 Adam A. Caparco 1 , Eric Pelletier 2 , Jean Louis Petit 2 , Aurélie Jouenne 2 , Bettina R. Bommarius 1 , Véronique Berardinis 2 , Anne Zaparucha 2 , Julie A. Champion 1 , Andreas S. Bommarius 1 , Carine Vergne‐Vaxelaire 2
Advanced Synthesis & Catalysis ( IF 5.4 ) Pub Date : 2020-03-30 , DOI: 10.1002/adsc.202000094 Adam A. Caparco 1 , Eric Pelletier 2 , Jean Louis Petit 2 , Aurélie Jouenne 2 , Bettina R. Bommarius 1 , Véronique Berardinis 2 , Anne Zaparucha 2 , Julie A. Champion 1 , Andreas S. Bommarius 1 , Carine Vergne‐Vaxelaire 2
Affiliation
|
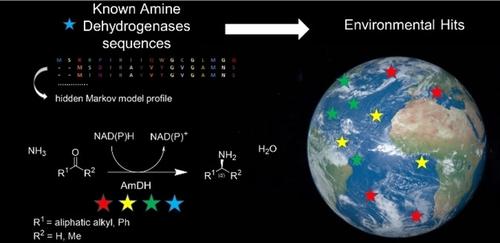
Amine dehydrogenases (AmDHs) catalyze the enzymatic reduction of ketones to amines, serving as a suitable biocatalytic route for amine synthesis. A limited number of experimentally validated native AmDHs (nat‐AmDHs) have been reported recently, expanding the sequences with this function to complement the small set of engineered enzymes. Since researchers can now probe into the vast diversity of enzymes within niche environments by a metagenomics approach, a tandem metagenomic and bioinformatic approach is a powerful tool to identify new members of limited enzyme families to access new features in an iterative fashion. The previously untapped biocatalytic reservoirs of the ocean environment and human microbiome were screened for potential AmDHs using a hidden Markov model. Among the hundreds of hits, a subset of 18 enzymes was selected for further characterization and were confirmed to display AmDH activity. Additional analysis on six enzymes confirmed altered cofactor specificities and variation in substrate scopes, catalytic efficiencies, and active site residues compared to the reference nat‐AmDHs previously described. Particularly, MATOUAmDH2 from an eukaryotic organism demonstrated specific activity of 11.07 and 0.88 U mg−1 toward isobutyraldehyde and 1,2‐cyclohexadione respectively. Their abundance among the screened environments was also described. The protein sequence diversity of validated AmDHs reached by this metagenomics mining strategy highlights the success of such an approach. Metagenomically mined proteins, including eukaryotic ones, stand to increase the reach of biocatalysis towards enviromentally benign processes.
中文翻译:

胺脱氢酶发现的超基因组挖掘
胺脱氢酶(AmDHs)催化酮类酶促还原为胺,这是胺合成的合适生物催化途径。最近报道了有限数量的经过实验验证的天然AmDHs(nat-AmDHs),从而扩展了具有此功能的序列以补充少量的工程化酶。由于研究人员现在可以通过宏基因组学方法探究利基环境中酶的多样性,因此串联宏基因组学和生物信息学方法是一种强大的工具,可用于识别有限酶家族的新成员以迭代方式访问新功能。使用隐藏的马尔可夫模型筛选了先前尚未开发的海洋环境和人类微生物组的生物催化储库中潜在的AmDH。在数百个热门歌曲中,选择了18种酶的一个子集进行进一步表征,并确认显示出AmDH活性。与先前描述的参比nat-AmDHs相比,对六种酶的进一步分析证实了辅因子的特异性改变以及底物范围,催化效率和活性位点残基的变化。特别地,来自真核生物的MATOUAmDH2表现出11.07和0.88U mg的比活性。-1对异丁醛和1,2-环己二酮。还描述了它们在筛选环境中的丰度。通过宏基因组学挖掘策略获得的经过验证的AmDHs的蛋白质序列多样性凸显了这种方法的成功。从基因组学角度开采的蛋白质,包括真核蛋白质,有望增加对环境良性过程的生物催化作用。
更新日期:2020-03-30
中文翻译:

胺脱氢酶发现的超基因组挖掘
胺脱氢酶(AmDHs)催化酮类酶促还原为胺,这是胺合成的合适生物催化途径。最近报道了有限数量的经过实验验证的天然AmDHs(nat-AmDHs),从而扩展了具有此功能的序列以补充少量的工程化酶。由于研究人员现在可以通过宏基因组学方法探究利基环境中酶的多样性,因此串联宏基因组学和生物信息学方法是一种强大的工具,可用于识别有限酶家族的新成员以迭代方式访问新功能。使用隐藏的马尔可夫模型筛选了先前尚未开发的海洋环境和人类微生物组的生物催化储库中潜在的AmDH。在数百个热门歌曲中,选择了18种酶的一个子集进行进一步表征,并确认显示出AmDH活性。与先前描述的参比nat-AmDHs相比,对六种酶的进一步分析证实了辅因子的特异性改变以及底物范围,催化效率和活性位点残基的变化。特别地,来自真核生物的MATOUAmDH2表现出11.07和0.88U mg的比活性。-1对异丁醛和1,2-环己二酮。还描述了它们在筛选环境中的丰度。通过宏基因组学挖掘策略获得的经过验证的AmDHs的蛋白质序列多样性凸显了这种方法的成功。从基因组学角度开采的蛋白质,包括真核蛋白质,有望增加对环境良性过程的生物催化作用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号