Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Markov State Model of Lassa Virus Nucleoprotein Reveals Large Structural Changes during the Trimer to Monomer Transition.
Structure ( IF 4.4 ) Pub Date : 2020-03-31 , DOI: 10.1016/j.str.2020.03.002 Jason G Pattis 1 , Eric R May 1
Structure ( IF 4.4 ) Pub Date : 2020-03-31 , DOI: 10.1016/j.str.2020.03.002 Jason G Pattis 1 , Eric R May 1
Affiliation
|
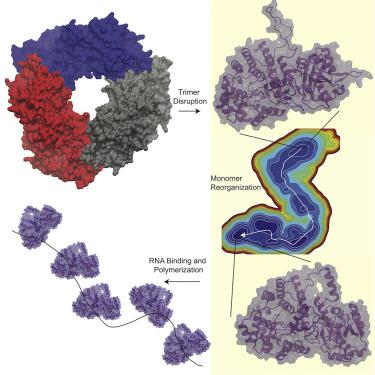
Lassa virus contains a nucleoprotein (NP) that encapsulates the viral genomic RNA forming the ribonucleoprotein (RNP). The NP forms trimers that do not bind RNA, but a structure of only the NP N-terminal domain was co-crystallized with RNA bound. These structures suggested a model in which the NP forms a trimer to keep the RNA gate closed, but then is triggered to undergo a change to a form competent for RNA binding. Here, we investigate the scenario in which the trimer is disrupted to observe whether monomeric NP undergoes significant conformational changes. From multi-microsecond molecular dynamics simulations and an adaptive sampling scheme to sample the conformational space, a Markov state model (MSM) is constructed. The MSM reveals an energetically favorable conformational change, with the most significant changes occurring at the domain interface. These results support a model in which significant structural reorganization of the NP is required for RNP formation.
中文翻译:

拉萨病毒核蛋白的马尔可夫状态模型揭示了三聚体到单体转变过程中的大结构变化。
拉萨病毒包含一个核蛋白(NP),该蛋白包封形成核糖核蛋白(RNP)的病毒基因组RNA。NP形成不结合RNA的三聚体,但是仅NP N末端结构域的结构与结合的RNA共结晶。这些结构提出了一个模型,其中NP形成三聚体以保持RNA门关闭,但随后被触发进行改变,转变为可与RNA结合的形式。在这里,我们调查三聚体被破坏的情况,以观察单体NP是否经历显着的构象变化。通过多微秒分子动力学模拟和自适应采样方案对构象空间进行采样,构建了马尔可夫状态模型(MSM)。MSM揭示了能量上有利的构象变化,最重要的变化发生在域界面上。这些结果支持一个模型,其中NP的形成需要NP的显着结构重组。
更新日期:2020-03-31
中文翻译:

拉萨病毒核蛋白的马尔可夫状态模型揭示了三聚体到单体转变过程中的大结构变化。
拉萨病毒包含一个核蛋白(NP),该蛋白包封形成核糖核蛋白(RNP)的病毒基因组RNA。NP形成不结合RNA的三聚体,但是仅NP N末端结构域的结构与结合的RNA共结晶。这些结构提出了一个模型,其中NP形成三聚体以保持RNA门关闭,但随后被触发进行改变,转变为可与RNA结合的形式。在这里,我们调查三聚体被破坏的情况,以观察单体NP是否经历显着的构象变化。通过多微秒分子动力学模拟和自适应采样方案对构象空间进行采样,构建了马尔可夫状态模型(MSM)。MSM揭示了能量上有利的构象变化,最重要的变化发生在域界面上。这些结果支持一个模型,其中NP的形成需要NP的显着结构重组。










































 京公网安备 11010802027423号
京公网安备 11010802027423号