当前位置:
X-MOL 学术
›
PLOS Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
DNA methylation and cis-regulation of gene expression by prostate cancer risk SNPs.
PLOS Genetics ( IF 4.0 ) Pub Date : 2020-03-30 , DOI: 10.1371/journal.pgen.1008667 James Y Dai 1, 2 , Xiaoyu Wang 1 , Bo Wang 3 , Wei Sun 1, 2 , Kristina M Jordahl 1 , Suzanne Kolb 1 , Yaw A Nyame 1, 4 , Jonathan L Wright 1, 4 , Elaine A Ostrander 5 , Ziding Feng 1, 2 , Janet L Stanford 1, 6
PLOS Genetics ( IF 4.0 ) Pub Date : 2020-03-30 , DOI: 10.1371/journal.pgen.1008667 James Y Dai 1, 2 , Xiaoyu Wang 1 , Bo Wang 3 , Wei Sun 1, 2 , Kristina M Jordahl 1 , Suzanne Kolb 1 , Yaw A Nyame 1, 4 , Jonathan L Wright 1, 4 , Elaine A Ostrander 5 , Ziding Feng 1, 2 , Janet L Stanford 1, 6
Affiliation
|
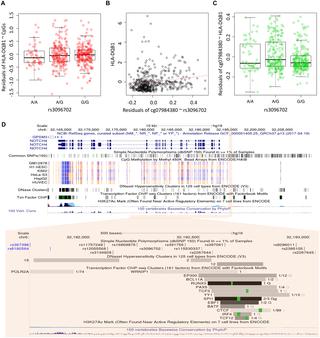
Genome-wide association studies have identified more than 100 SNPs that increase the risk of prostate cancer (PrCa). We identify and compare expression quantitative trait loci (eQTLs) and CpG methylation quantitative trait loci (meQTLs) among 147 established PrCa risk SNPs in primary prostate tumors (n = 355 from a Seattle-based study and n = 495 from The Cancer Genome Atlas, TCGA) and tumor-adjacent, histologically benign samples (n = 471 from a Mayo Clinic study). The role of DNA methylation in eQTL regulation of gene expression was investigated by data triangulation using several causal inference approaches, including a proposed adaptation of the Causal Inference Test (CIT) for causal direction. Comparing eQTLs between tumors and benign samples, we show that 98 of the 147 risk SNPs were identified as eQTLs in the tumor-adjacent benign samples, and almost all 34 eQTL identified in tumor sets were also eQTLs in the benign samples. Three lines of results support the causal role of DNA methylation. First, nearly 100 of the 147 risk SNPs were identified as meQTLs in one tumor set, and almost all eQTLs in tumors were meQTLs. Second, the loss of eQTLs in tumors relative to benign samples was associated with altered DNA methylation. Third, among risk SNPs identified as both eQTLs and meQTLs, mediation analyses suggest that over two-thirds have evidence of a causal role for DNA methylation, mostly mediating genetic influence on gene expression. In summary, we provide a comprehensive catalog of eQTLs, meQTLs and putative cancer genes for known PrCa risk SNPs. We observe that a substantial portion of germline eQTL regulatory mechanisms are maintained in the tumor development, despite somatic alterations in tumor genome. Finally, our mediation analyses illuminate the likely intermediary role of CpG methylation in eQTL regulation of gene expression.
中文翻译:

DNA 甲基化和前列腺癌风险 SNP 对基因表达的顺式调节。
全基因组关联研究已发现 100 多个 SNP 会增加前列腺癌 (PrCa) 的风险。我们在原发性前列腺肿瘤中 147 个已确定的 PrCa 风险 SNP 中鉴定并比较了表达数量性状位点 (eQTL) 和 CpG 甲基化数量性状位点 (meQTL)(n = 355 来自西雅图的研究,n = 495 来自癌症基因组图谱, TCGA)和肿瘤邻近的组织学良性样本(n = 471,来自 Mayo Clinic 研究)。 DNA 甲基化在 eQTL 基因表达调控中的作用通过数据三角测量使用几种因果推理方法进行了研究,包括对因果方向的因果推理测试 (CIT) 的拟议修改。比较肿瘤和良性样本之间的 eQTL,我们发现 147 个风险 SNP 中的 98 个被识别为肿瘤相邻良性样本中的 eQTL,并且在肿瘤集中识别的几乎所有 34 个 eQTL 也是良性样本中的 eQTL。三行结果支持 DNA 甲基化的因果作用。首先,147 个风险 SNP 中的近 100 个在一组肿瘤中被鉴定为 meQTL,并且肿瘤中几乎所有 eQTL 都是 meQTL。其次,相对于良性样本,肿瘤中 eQTL 的丢失与 DNA 甲基化的改变有关。第三,在被确定为 eQTL 和 meQTL 的风险 SNP 中,中介分析表明,超过三分之二的 SNP 具有 DNA 甲基化因果作用的证据,主要介导对基因表达的遗传影响。总之,我们为已知的 PrCa 风险 SNP 提供了 eQTL、meQTL 和推定癌症基因的综合目录。我们观察到,尽管肿瘤基因组发生体细胞改变,但生殖系 eQTL 调控机制的很大一部分在肿瘤发展过程中得以维持。 最后,我们的中介分析阐明了 CpG 甲基化在 eQTL 基因表达调控中可能发挥的中介作用。
更新日期:2020-04-06
中文翻译:

DNA 甲基化和前列腺癌风险 SNP 对基因表达的顺式调节。
全基因组关联研究已发现 100 多个 SNP 会增加前列腺癌 (PrCa) 的风险。我们在原发性前列腺肿瘤中 147 个已确定的 PrCa 风险 SNP 中鉴定并比较了表达数量性状位点 (eQTL) 和 CpG 甲基化数量性状位点 (meQTL)(n = 355 来自西雅图的研究,n = 495 来自癌症基因组图谱, TCGA)和肿瘤邻近的组织学良性样本(n = 471,来自 Mayo Clinic 研究)。 DNA 甲基化在 eQTL 基因表达调控中的作用通过数据三角测量使用几种因果推理方法进行了研究,包括对因果方向的因果推理测试 (CIT) 的拟议修改。比较肿瘤和良性样本之间的 eQTL,我们发现 147 个风险 SNP 中的 98 个被识别为肿瘤相邻良性样本中的 eQTL,并且在肿瘤集中识别的几乎所有 34 个 eQTL 也是良性样本中的 eQTL。三行结果支持 DNA 甲基化的因果作用。首先,147 个风险 SNP 中的近 100 个在一组肿瘤中被鉴定为 meQTL,并且肿瘤中几乎所有 eQTL 都是 meQTL。其次,相对于良性样本,肿瘤中 eQTL 的丢失与 DNA 甲基化的改变有关。第三,在被确定为 eQTL 和 meQTL 的风险 SNP 中,中介分析表明,超过三分之二的 SNP 具有 DNA 甲基化因果作用的证据,主要介导对基因表达的遗传影响。总之,我们为已知的 PrCa 风险 SNP 提供了 eQTL、meQTL 和推定癌症基因的综合目录。我们观察到,尽管肿瘤基因组发生体细胞改变,但生殖系 eQTL 调控机制的很大一部分在肿瘤发展过程中得以维持。 最后,我们的中介分析阐明了 CpG 甲基化在 eQTL 基因表达调控中可能发挥的中介作用。









































 京公网安备 11010802027423号
京公网安备 11010802027423号