Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Qualitative Ras pathway signature for cetuximab therapy reveals resistant mechanism in colorectal cancer.
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-03-25 , DOI: 10.1111/febs.15306 Kai Song 1 , Haibo Lu 2 , Liangliang Jin 1 , Kai Wang 1 , Wenbing Guo 1 , Hailong Zheng 1 , Keru Li 1 , Chuchu He 2 , Tianyi You 1 , Yelin Fu 1 , Jing Yang 1 , Wenyuan Zhao 1 , Zheng Guo 1
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-03-25 , DOI: 10.1111/febs.15306 Kai Song 1 , Haibo Lu 2 , Liangliang Jin 1 , Kai Wang 1 , Wenbing Guo 1 , Hailong Zheng 1 , Keru Li 1 , Chuchu He 2 , Tianyi You 1 , Yelin Fu 1 , Jing Yang 1 , Wenyuan Zhao 1 , Zheng Guo 1
Affiliation
|
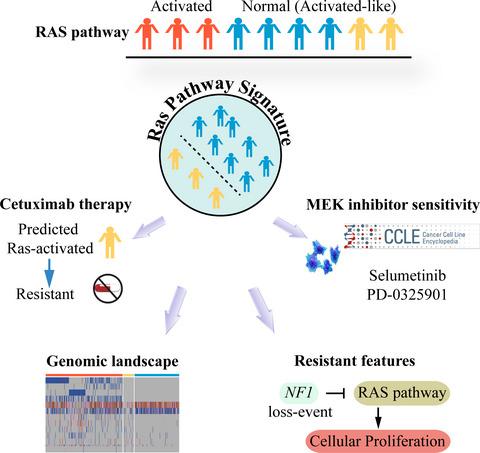
Cetuximab therapy, which heavily relies on the activation of Ras pathway, has been used in KRAS, NRAS, BRAF, and PIK3CA wild‐type colorectal cancer (CRC) (Ras‐normal). However, the response rate only reached 60%, due to false‐negative mutation detection and mutation‐like transcriptome features in wild‐type patients. Herein, by integrating RNA‐seq, microarray, and mutation data, we developed a Ras pathway signature by characterizing KRAS/NRAS/BRAF/PIK3CA mutations to identify the hidden nonresponders from the Ras‐normal patients by mutation detection. Using public and in‐house data of CRC patients treated with cetuximab, discovery of the signature could identify cetuximab‐resistant samples from the Ras‐normal samples. Cetuximab resistance‐related genes, such as PTEN, were significantly and frequently mutated in the identified Ras‐activated samples, whereas two cetuximab sensitivity‐related genes, APC and TP53, showed comutation and significantly higher mutation frequencies in the remaining Ras‐normal samples. Furthermore, all the NF1‐ and BCL2L1‐mutated samples were identified as Ras‐activated from the Ras‐normal samples by the Ras pathway signature with significantly under‐regulated expression. Genes co‐expressed with the two genes were both involved in Ras signaling pathway, the out‐of‐control of which could be attributed by the genes' loss‐of‐function mutations. To improve the treatment of cetuximab in CRC, NF1 and BCL2L1 could be used as complementary detection technique to those applied in clinical. In conclusion, the proposed Ras pathway signature could identify the hidden CRC patients resistant to cetuximab therapy and help to reveal resistance mechanisms.
中文翻译:

西妥昔单抗治疗的定性Ras通路特征揭示了结直肠癌的耐药机制。
高度依赖Ras途径活化的西妥昔单抗疗法已用于KRAS,NRAS,BRAF和PIK3CA野生型结直肠癌(CRC)(Rasnormal)。但是,由于假阴性突变检测和野生型患者的突变样转录组特征,反应率仅达到60%。本文中,通过整合RNA-seq,微阵列和突变数据,我们通过表征KRAS / NRAS / BRAF / PIK3CA形成了Ras途径签名。通过突变检测来识别Ras正常患者中隐藏的无反应者。利用西妥昔单抗治疗的CRC患者的公开和内部数据,发现特征可以从Ras正常样本中识别出耐西妥昔单抗的样本。在已鉴定的Ras激活样品中,西妥昔单抗抗性相关基因(如PTEN)显着且频繁地突变,而在其余Ras正常样品中,两个与西妥昔单抗敏感性相关的基因APC和TP53表现出易位和显着更高的突变频率。此外,所有NF1-和BCL2L1-突变的样品被Ras途径签名从Ras正常样品中鉴定为Ras激活,且表达水平明显低于正常水平。与这两个基因共表达的基因都参与了Ras信号通路,其失控可能是由于基因的功能丧失突变所致。为了改善西妥昔单抗在CRC中的治疗,可以将NF1和BCL2L1用作对临床应用的补充检测技术。总之,拟议的Ras通路签名可以识别出对西妥昔单抗治疗耐药的隐匿性CRC患者,并有助于揭示耐药机制。
更新日期:2020-03-25
中文翻译:

西妥昔单抗治疗的定性Ras通路特征揭示了结直肠癌的耐药机制。
高度依赖Ras途径活化的西妥昔单抗疗法已用于KRAS,NRAS,BRAF和PIK3CA野生型结直肠癌(CRC)(Rasnormal)。但是,由于假阴性突变检测和野生型患者的突变样转录组特征,反应率仅达到60%。本文中,通过整合RNA-seq,微阵列和突变数据,我们通过表征KRAS / NRAS / BRAF / PIK3CA形成了Ras途径签名。通过突变检测来识别Ras正常患者中隐藏的无反应者。利用西妥昔单抗治疗的CRC患者的公开和内部数据,发现特征可以从Ras正常样本中识别出耐西妥昔单抗的样本。在已鉴定的Ras激活样品中,西妥昔单抗抗性相关基因(如PTEN)显着且频繁地突变,而在其余Ras正常样品中,两个与西妥昔单抗敏感性相关的基因APC和TP53表现出易位和显着更高的突变频率。此外,所有NF1-和BCL2L1-突变的样品被Ras途径签名从Ras正常样品中鉴定为Ras激活,且表达水平明显低于正常水平。与这两个基因共表达的基因都参与了Ras信号通路,其失控可能是由于基因的功能丧失突变所致。为了改善西妥昔单抗在CRC中的治疗,可以将NF1和BCL2L1用作对临床应用的补充检测技术。总之,拟议的Ras通路签名可以识别出对西妥昔单抗治疗耐药的隐匿性CRC患者,并有助于揭示耐药机制。











































 京公网安备 11010802027423号
京公网安备 11010802027423号