当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Programming the Sequential Release of DNA.
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-04-08 , DOI: 10.1021/acssynbio.9b00398 Dominic Scalise , Moshe Rubanov , Katherine Miller , Leo Potters , Madeline Noble , Rebecca Schulman
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-04-08 , DOI: 10.1021/acssynbio.9b00398 Dominic Scalise , Moshe Rubanov , Katherine Miller , Leo Potters , Madeline Noble , Rebecca Schulman
|
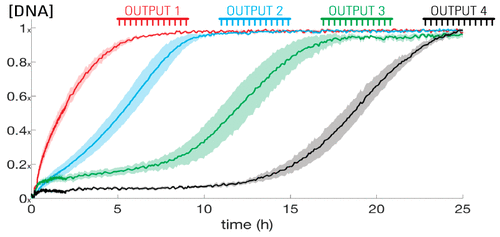
This study presents a mechanism for releasing a series of different short DNA sequences from sequestered complexes, one after another, using coupled biochemical reactions. The process uses stages of coupled DNA strand-displacement reactions that first release an output molecule and then trigger the initiation of the next release stage. We demonstrate the sequential release of 25 nM of four different sequences of DNA over a day, both with and without a centralized "clock" mechanism to regulate release timing. We then demonstrate how the presence of a target input molecule can determine which of several different release pathways are activated, analogous to branching conditional statements in computer programming. This sequential release circuit offers a means to schedule downstream chemical events, such as steps in the assembly of a nanostructure, or stages in a material's response to a stimulus.
中文翻译:

对DNA的顺序释放进行编程。
这项研究提出了一种利用偶联的生化反应从螯合复合物中释放一系列不同的短DNA序列的机制。该过程使用耦合的DNA链置换反应阶段,该阶段首先释放输出分子,然后触发下一个释放阶段的启动。我们演示了一天中有四个DNA序列的25 nM的顺序释放,无论有没有中央的“时钟”机制来调节释放时间。然后,我们演示了目标输入分子的存在如何确定几种不同的释放途径中的哪一种,类似于计算机编程中的分支条件语句。该顺序释放回路提供了一种安排下游化学事件的方法,
更新日期:2020-04-23
中文翻译:

对DNA的顺序释放进行编程。
这项研究提出了一种利用偶联的生化反应从螯合复合物中释放一系列不同的短DNA序列的机制。该过程使用耦合的DNA链置换反应阶段,该阶段首先释放输出分子,然后触发下一个释放阶段的启动。我们演示了一天中有四个DNA序列的25 nM的顺序释放,无论有没有中央的“时钟”机制来调节释放时间。然后,我们演示了目标输入分子的存在如何确定几种不同的释放途径中的哪一种,类似于计算机编程中的分支条件语句。该顺序释放回路提供了一种安排下游化学事件的方法,











































 京公网安备 11010802027423号
京公网安备 11010802027423号