当前位置:
X-MOL 学术
›
Cancer Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Molecular natural history of breast cancer: Leveraging transcriptomics to predict breast cancer progression and aggressiveness.
Cancer Medicine ( IF 2.9 ) Pub Date : 2020-03-23 , DOI: 10.1002/cam4.2996 Daniel J Cook 1, 2 , Jonatan Kallus 3 , Rebecka Jörnsten 3 , Jens Nielsen 1, 2, 4, 5
Cancer Medicine ( IF 2.9 ) Pub Date : 2020-03-23 , DOI: 10.1002/cam4.2996 Daniel J Cook 1, 2 , Jonatan Kallus 3 , Rebecka Jörnsten 3 , Jens Nielsen 1, 2, 4, 5
Affiliation
|
BACKGROUND
Characterizing breast cancer progression and aggressiveness relies on categorical descriptions of tumor stage and grade. Interpreting these categorical descriptions is challenging because stage convolutes the size and spread of the tumor and no consensus exists to define high/low grade tumors.
METHODS
We address this challenge of heterogeneity in patient-specific cancer samples by adapting and applying several tools originally created for understanding heterogeneity and phenotype development in single cells (specifically, single-cell topological data analysis and Wanderlust) to create a continuous metric describing breast cancer progression using bulk RNA-seq samples from individual patient tumors. We also created a linear regression-based method to predict tumor aggressiveness in vivo from bulk RNA-seq data.
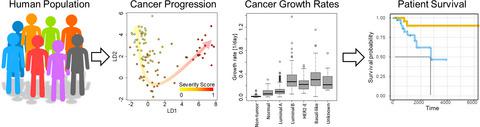
RESULTS
We found that breast cancer proceeds along three convergent phenotype trajectories: luminal, HER2-enriched, and basal-like. Furthermore, 31 296 genes (for luminal cancers), 17 827 genes (for HER2-enriched), and 18 505 genes (for basal-like) are dynamically differentially expressed during breast cancer progression. Across progression trajectories, our results show that expression of genes related to ADP-ribosylation decreased as tumors progressed (while PARP1 and PARP2 increased or remained stable), suggesting the potential for a differential response to PARP inhibitors based on cancer progression. Additionally, we developed a 132-gene expression regression equation to predict mitotic index and a 23-gene expression regression equation to predict growth rate from a single breast cancer biopsy.
CONCLUSION
Our results suggest that breast cancer dynamically changes during disease progression, and growth rate of the cancer cells is associated with distinct transcriptional profiles.
中文翻译:

乳腺癌的分子自然史:利用转录组学预测乳腺癌的进展和侵袭性。
背景描述乳腺癌进展和侵袭性的特征依赖于肿瘤分期和分级的分类描述。解释这些分类描述具有挑战性,因为分期使肿瘤的大小和扩散变得复杂,并且对于高/低级别肿瘤的定义尚无共识。方法我们通过调整和应用最初为理解单细胞异质性和表型发展而创建的几种工具(特别是单细胞拓扑数据分析和 Wanderlust)来创建描述乳腺癌的连续指标,从而解决患者特定癌症样本中异质性的挑战使用来自个体患者肿瘤的大量 RNA-seq 样本进行进展。我们还创建了一种基于线性回归的方法,根据大量 RNA-seq 数据预测体内肿瘤侵袭性。结果我们发现乳腺癌沿着三种趋同的表型轨迹发展:管腔型、HER2 富集型和基底样型。此外,31 296 个基因(管腔癌)、17 827 个基因(HER2 富集)和 18 505 个基因(基底样)在乳腺癌进展过程中动态差异表达。在整个进展轨迹中,我们的结果表明,与 ADP-核糖基化相关的基因表达随着肿瘤的进展而减少(而 PARP1 和 PARP2 增加或保持稳定),这表明根据癌症进展,对 PARP 抑制剂的反应可能存在差异。此外,我们还开发了一个 132 基因表达回归方程来预测有丝分裂指数,以及一个 23 基因表达回归方程来预测单个乳腺癌活检的生长率。 结论我们的结果表明乳腺癌在疾病进展过程中动态变化,并且癌细胞的生长速率与不同的转录谱相关。
更新日期:2020-03-23
中文翻译:

乳腺癌的分子自然史:利用转录组学预测乳腺癌的进展和侵袭性。
背景描述乳腺癌进展和侵袭性的特征依赖于肿瘤分期和分级的分类描述。解释这些分类描述具有挑战性,因为分期使肿瘤的大小和扩散变得复杂,并且对于高/低级别肿瘤的定义尚无共识。方法我们通过调整和应用最初为理解单细胞异质性和表型发展而创建的几种工具(特别是单细胞拓扑数据分析和 Wanderlust)来创建描述乳腺癌的连续指标,从而解决患者特定癌症样本中异质性的挑战使用来自个体患者肿瘤的大量 RNA-seq 样本进行进展。我们还创建了一种基于线性回归的方法,根据大量 RNA-seq 数据预测体内肿瘤侵袭性。结果我们发现乳腺癌沿着三种趋同的表型轨迹发展:管腔型、HER2 富集型和基底样型。此外,31 296 个基因(管腔癌)、17 827 个基因(HER2 富集)和 18 505 个基因(基底样)在乳腺癌进展过程中动态差异表达。在整个进展轨迹中,我们的结果表明,与 ADP-核糖基化相关的基因表达随着肿瘤的进展而减少(而 PARP1 和 PARP2 增加或保持稳定),这表明根据癌症进展,对 PARP 抑制剂的反应可能存在差异。此外,我们还开发了一个 132 基因表达回归方程来预测有丝分裂指数,以及一个 23 基因表达回归方程来预测单个乳腺癌活检的生长率。 结论我们的结果表明乳腺癌在疾病进展过程中动态变化,并且癌细胞的生长速率与不同的转录谱相关。









































 京公网安备 11010802027423号
京公网安备 11010802027423号