Nature Communications ( IF 14.7 ) Pub Date : 2020-03-25 , DOI: 10.1038/s41467-020-15346-1 Brian C Searle 1, 2 , Kristian E Swearingen 1 , Christopher A Barnes 3 , Tobias Schmidt 4 , Siegfried Gessulat 4, 5 , Bernhard Küster 4, 6 , Mathias Wilhelm 4
|
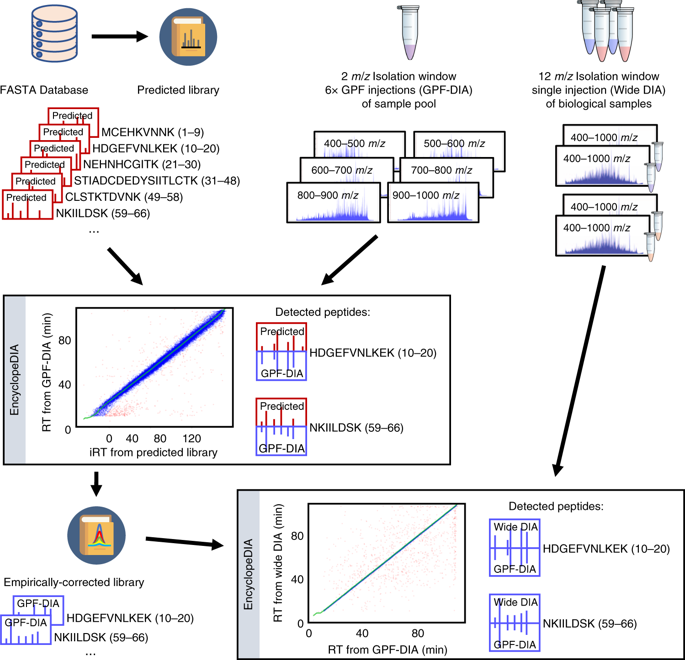
Data-independent acquisition approaches typically rely on experiment-specific spectrum libraries, requiring offline fractionation and tens to hundreds of injections. We demonstrate a library generation workflow that leverages fragmentation and retention time prediction to build libraries containing every peptide in a proteome, and then refines those libraries with empirical data. Our method specifically enables rapid, experiment-specific library generation for non-model organisms, which we demonstrate using the malaria parasite Plasmodium falciparum, and non-canonical databases, which we show by detecting missense variants in HeLa.
中文翻译:

为 DIA MS 生成高质量的文库,并根据经验校正肽预测。
与数据无关的采集方法通常依赖于特定于实验的谱库,需要离线分馏和数十到数百次进样。我们演示了一个文库生成工作流程,该工作流程利用片段化和保留时间预测来构建包含蛋白质组中每个肽的文库,然后用经验数据完善这些文库。我们的方法特别能够为非模型生物体快速生成特定于实验的文库,我们使用疟原虫恶性疟原虫证明了这一点,以及非规范数据库,我们通过检测 HeLa 中的错义变异来证明这一点。











































 京公网安备 11010802027423号
京公网安备 11010802027423号