Molecular Psychiatry ( IF 9.6 ) Pub Date : 2020-03-25 , DOI: 10.1038/s41380-020-0715-7 Shiwu Li 1, 2 , Yifan Li 1, 2 , Xiaoyan Li 1, 2 , Jiewei Liu 1 , Yongxia Huo 1 , Junyang Wang 1, 2 , Zhongchun Liu 3 , Ming Li 1, 2 , Xiong-Jian Luo 1, 2, 4, 5
|
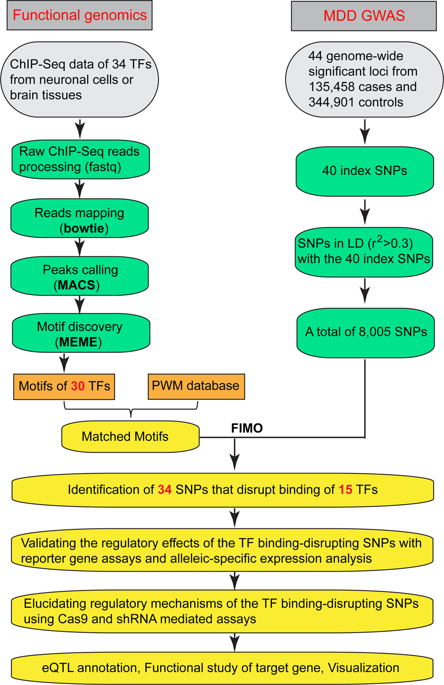
Major depressive disorder (MDD) is one of the most prevalent psychiatric disorders and a leading cause of disability worldwide. Though recent genome-wide association studies (GWAS) have identified multiple risk variants for MDD, how these variants confer MDD risk remains largely unknown. Here we systematically characterize the regulatory mechanism of MDD risk variants using a functional genomics approach. By integrating chromatin immunoprecipitation sequencing (ChIP-Seq) (from human brain tissues or neuronal cells) and position weight matrix (PWM) data, we identified 34 MDD risk SNPs that disrupt the binding of 15 transcription factors (TFs). We verified the regulatory effect of the TF binding–disrupting SNPs with reporter gene assays, allelic-specific expression analysis, and CRISPR-Cas9-mediated genome editing. Expression quantitative trait loci (eQTL) analysis identified the target genes that might be regulated by these regulatory risk SNPs. Finally, we found that NEGR1 (regulated by the TF binding–disrupting MDD risk SNP rs3101339) was dysregulated in the brains of MDD cases compared with controls, implying that rs3101339 may confer MDD risk by affecting NEGR1 expression. Our findings reveal how genetic variants contribute to MDD risk by affecting TF binding and gene regulation. More importantly, our study identifies the potential MDD causal variants and their target genes, thus providing pivotal candidates for future mechanistic study and drug development.
中文翻译:

重度抑郁症风险变异的调节机制。
重度抑郁症 (MDD) 是全球最普遍的精神疾病之一,也是导致残疾的主要原因。尽管最近的全基因组关联研究 (GWAS) 已经确定了 MDD 的多种风险变异,但这些变异如何赋予 MDD 风险仍然很大程度上未知。在这里,我们使用功能基因组学方法系统地描述了 MDD 风险变异的调节机制。通过整合染色质免疫沉淀测序 (ChIP-Seq)(来自人脑组织或神经元细胞)和位置权重矩阵 (PWM) 数据,我们确定了 34 个破坏 15 种转录因子 (TF) 结合的 MDD 风险 SNP。我们通过报告基因检测、等位基因特异性表达分析和 CRISPR-Cas9 介导的基因组编辑验证了 TF 结合破坏 SNP 的调节作用。表达数量性状基因座 (eQTL) 分析确定了可能受这些调控风险 SNP 调控的靶基因。最后,我们发现与对照组相比, NEGR1(受 TF 结合破坏 MDD 风险 SNP rs3101339 调节)在 MDD 病例的大脑中失调,这意味着 rs3101339 可能通过影响NEGR1表达来赋予 MDD 风险。我们的研究结果揭示了遗传变异如何通过影响 TF 结合和基因调控来增加 MDD 风险。更重要的是,我们的研究确定了潜在的 MDD 因果变异及其靶基因,从而为未来的机制研究和药物开发提供了关键的候选者。











































 京公网安备 11010802027423号
京公网安备 11010802027423号