Nature ( IF 50.5 ) Pub Date : 2020-03-25 , DOI: 10.1038/s41586-020-2125-z Samuel Collombet 1, 2 , Noémie Ranisavljevic 1, 3 , Takashi Nagano 4, 5 , Csilla Varnai 4, 6 , Tarak Shisode 7 , Wing Leung 4, 5 , Tristan Piolot 1 , Rafael Galupa 1, 2 , Maud Borensztein 1 , Nicolas Servant 8 , Peter Fraser 4, 9 , Katia Ancelin 1 , Edith Heard 1, 2
|
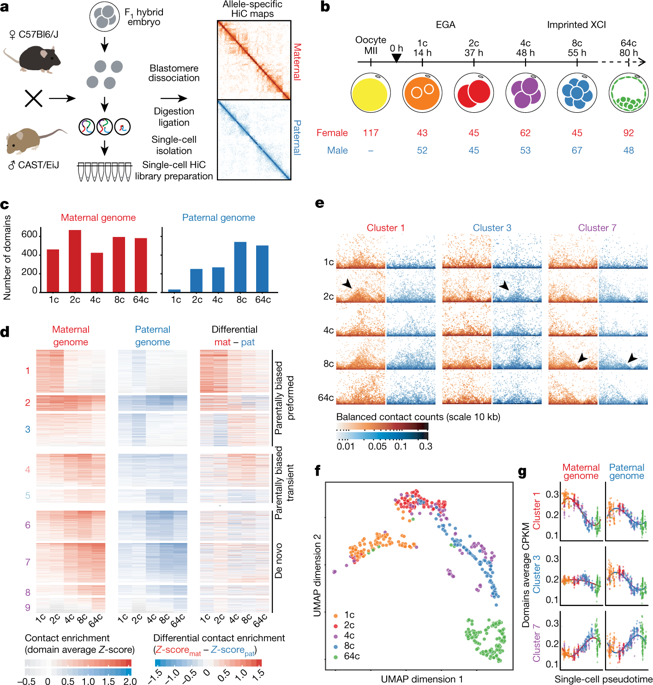
Paternal and maternal epigenomes undergo marked changes after fertilization1. Recent epigenomic studies have revealed the unusual chromatin landscapes that are present in oocytes, sperm and early preimplantation embryos, including atypical patterns of histone modifications2,3,4 and differences in chromosome organization and accessibility, both in gametes5,6,7,8 and after fertilization5,8,9,10. However, these studies have led to very different conclusions: the global absence of local topological-associated domains (TADs) in gametes and their appearance in the embryo8,9 versus the pre-existence of TADs and loops in the zygote5,11. The questions of whether parental structures can be inherited in the newly formed embryo and how these structures might relate to allele-specific gene regulation remain open. Here we map genomic interactions for each parental genome (including the X chromosome), using an optimized single-cell high-throughput chromosome conformation capture (HiC) protocol12,13, during preimplantation in the mouse. We integrate chromosome organization with allelic expression states and chromatin marks, and reveal that higher-order chromatin structure after fertilization coincides with an allele-specific enrichment of methylation of histone H3 at lysine 27. These early parental-specific domains correlate with gene repression and participate in parentally biased gene expression—including in recently described, transiently imprinted loci14. We also find TADs that arise in a non-parental-specific manner during a second wave of genome assembly. These de novo domains are associated with active chromatin. Finally, we obtain insights into the relationship between TADs and gene expression by investigating structural changes to the paternal X chromosome before and during X chromosome inactivation in preimplantation female embryos15. We find that TADs are lost as genes become silenced on the paternal X chromosome but linger in regions that escape X chromosome inactivation. These findings demonstrate the complex dynamics of three-dimensional genome organization and gene expression during early development.
中文翻译:

早期胚胎发生中染色体组织的亲代胚胎转换
父系和母系表观基因组在受精1后发生显着变化。最近的表观基因组学研究揭示了卵母细胞、精子和早期植入前胚胎中存在的不寻常的染色质景观,包括非典型的组蛋白修饰模式2,3,4以及染色体组织和可及性的差异,均在配子中5,6,7,8和受精后5,8,9,10。然而,这些研究得出了截然不同的结论:配子中局部拓扑相关域 (TAD) 的全球缺失及其在胚胎中的出现8,9与受精卵中 TAD 和环的预先存在5,11. 亲本结构是否可以在新形成的胚胎中遗传以及这些结构如何与等位基因特异性基因调控相关的问题仍然悬而未决。在这里,我们使用优化的单细胞高通量染色体构象捕获 (HiC) 协议12,13绘制每个亲本基因组(包括 X 染色体)的基因组相互作用图,在小鼠植入前。我们将染色体组织与等位基因表达状态和染色质标记整合在一起,并揭示受精后的高级染色质结构与组蛋白 H3 在赖氨酸 27 处甲基化的等位基因特异性富集一致。这些早期的亲本特异性结构域与基因抑制相关并参与在亲本偏倚的基因表达中——包括最近描述的瞬时印迹基因座14. 我们还发现在第二波基因组组装过程中以非亲本特异性方式出现的 TAD。这些从头结构域与活性染色质相关。最后,我们通过调查在植入前雌性胚胎中 X 染色体失活15之前和期间父亲 X 染色体的结构变化,获得了对 TAD 和基因表达之间关系的见解。我们发现随着基因在父系 X 染色体上变得沉默,TAD 会丢失,但会在逃脱 X 染色体失活的区域徘徊。这些发现证明了早期发育过程中三维基因组组织和基因表达的复杂动态。











































 京公网安备 11010802027423号
京公网安备 11010802027423号