Nature Structural & Molecular Biology ( IF 16.8 ) Pub Date : 2020-03-23 , DOI: 10.1038/s41594-020-0398-4 Yulong Song 1 , Wenbing Yang 1 , Qiang Fu 1 , Liang Wu 2 , Xueni Zhao 1 , Yusen Zhang 1 , Rui Zhang 1, 3
|
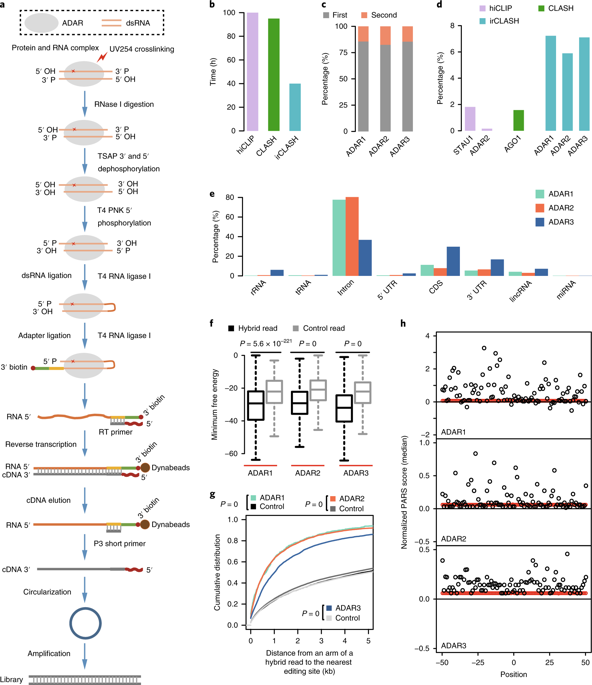
Adenosine deaminases acting on RNA (ADARs) convert adenosines to inosines in double-stranded RNA (dsRNA) in animals. Despite their importance, ADAR RNA substrates have not been mapped extensively in vivo. Here we develop irCLASH to map RNA substrates recognized by human ADARs and uncover features that determine their binding affinity and editing efficiency. We also observe a dominance of long-range interactions within ADAR substrates and analyze differences between ADAR1 and ADAR2 editing substrates. Moreover, we unexpectedly discovered that ADAR proteins bind dsRNA substrates tandemly in vivo, each with a 50-bp footprint. Using RNA duplexes recognized by ADARs as readout of pre-messenger RNA structures, we reveal distinct higher-order architectures between pre-messenger RNAs and mRNAs. Our transcriptome-wide atlas of ADAR substrates and the features governing RNA editing observed in our study will assist in the rational design of guide RNAs for ADAR-mediated RNA base editing.
中文翻译:

irCLASH揭示了人类ADAR识别的RNA底物。
作用于RNA(ADAR)的腺苷脱氨基酶将动物双链RNA(dsRNA)中的腺苷转化为肌苷。尽管具有重要意义,但ADAR RNA底物尚未在体内进行广泛定位。在这里,我们开发了irCLASH来绘制人类ADAR识别的RNA底物,并揭示决定其结合亲和力和编辑效率的特征。我们还观察到ADAR基质内远程相互作用的优势,并分析了ADAR1和ADAR2编辑基质之间的差异。此外,我们出乎意料地发现ADAR蛋白在体内串联结合dsRNA底物,每个底物都有50 bp的足迹。使用由ADARs识别的RNA双链体作为信使前RNA结构的读数,我们揭示了信使前RNA与mRNA之间独特的高阶结构。



























 京公网安备 11010802027423号
京公网安备 11010802027423号