The Journal of Antibiotics ( IF 2.1 ) Pub Date : 2020-03-24 , DOI: 10.1038/s41429-020-0296-3 Anne Leonard 1 , Kevin Möhlis 1 , Rabea Schlüter 2 , Edward Taylor 3 , Michael Lalk 1 , Karen Methling 1
|
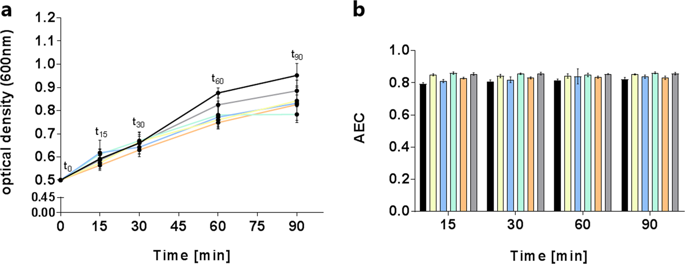
The Gram-positive bacterium Streptococcus pneumoniae is one of the common causes of community acquired pneumonia, meningitis, and otitis media. Analyzing the metabolic adaptation toward environmental stress conditions improves our understanding of its pathophysiology and its dependency on host-derived nutrients. In this study, extra- and intracellular metabolic profiles were evaluated to investigate the impact of antimicrobial compounds targeting different pathways of the metabolome of S. pneumoniae TIGR4Δcps. For the metabolomics approach, we analyzed the complex variety of metabolites by using 1H NMR, HPLC-MS, and GC–MS as different analytical techniques. Through this combination, we detected nearly 120 metabolites. For each antimicrobial compound, individual metabolic effects were detected that often comprised global biosynthetic pathways. Cefotaxime altered amino acids metabolism and carbon metabolism. The purine and pyrimidine metabolic pathways were mostly affected by moxifloxacin treatment. The combination of cefotaxime and azithromycin intensified the stress response compared with the use of the single antibiotic. However, we observed that three cell wall metabolites were altered only by treatment with the combination of the two antibiotics. Only moxifloxacin stress-induced alternation in CDP-ribitol concentration. Teixobactin-Arg10 resulted in global changes of pneumococcal metabolism. To meet the growing requirements for new antibiotics, our metabolomics approach has shown to be a promising complement to other OMICs investigations allowing insights into the mode of action of novel antimicrobial compounds.
中文翻译:

探索肺炎链球菌对抗生素的代谢适应性。
革兰氏阳性菌肺炎链球菌是引起社区获得性肺炎,脑膜炎和中耳炎的常见原因之一。分析代谢物对环境压力条件的适应性,可以增进我们对其病理生理学及其对宿主衍生养分的依赖性的了解。在这项研究中,细胞外和细胞内的代谢特征进行评估,以调查靶向的代谢组的不同途径的抗微生物化合物的影响肺炎链球菌TIGR4 Δcps。对于代谢组学方法,我们使用1来分析复杂的代谢产物H NMR,HPLC-MS和GC-MS作为不同的分析技术。通过这种组合,我们检测了近120种代谢物。对于每种抗微生物化合物,检测到的个体代谢效应通常包括整体生物合成途径。头孢噻肟改变了氨基酸代谢和碳代谢。嘌呤和嘧啶的代谢途径主要受莫西沙星治疗的影响。与使用单一抗生素相比,头孢噻肟和阿奇霉素的组合可增强应激反应。但是,我们观察到只有通过两种抗生素的组合治疗才能改变三种细胞壁代谢物。仅莫西沙星在CDP-核糖醇浓度上引起应激变化。Teixobactin-Arg10导致了肺炎球菌代谢的整体变化。











































 京公网安备 11010802027423号
京公网安备 11010802027423号