当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structural Analysis and Identification of False Positive Hits in Luciferase-Based Assays.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-03-23 , DOI: 10.1021/acs.jcim.9b01188 Zi-Yi Yang 1 , Jie Dong 2 , Zhi-Jiang Yang 1 , Ai-Ping Lu 3 , Ting-Jun Hou 4 , Dong-Sheng Cao 1, 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-03-23 , DOI: 10.1021/acs.jcim.9b01188 Zi-Yi Yang 1 , Jie Dong 2 , Zhi-Jiang Yang 1 , Ai-Ping Lu 3 , Ting-Jun Hou 4 , Dong-Sheng Cao 1, 3
Affiliation
|
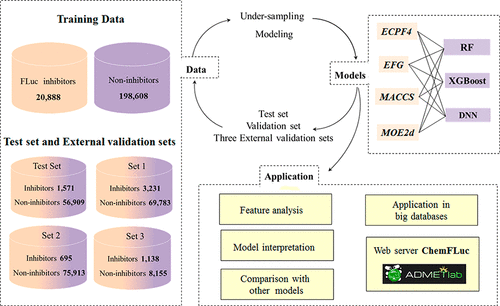
Luciferase-based bioluminescence detection techniques are highly favored in high-throughput screening (HTS), in which the firefly luciferase (FLuc) is the most commonly used variant. However, FLuc inhibitors can interfere with the activity of luciferase, which may result in false positive signals in HTS assays. In order to reduce the unnecessary cost of time and money, an in silico prediction model for FLuc inhibitors is highly desirable. In this study, we built an extensive data set consisting of 20 888 FLuc inhibitors and 198 608 noninhibitors, and then developed a group of classification models based on the combination of three machine learning (ML) algorithms and four types of molecular representations. The best prediction model based on XGBoost and ECFP4 and MOE2d descriptors yielded a balanced accuracy (BA) of 0.878 and an area under the receiver operating characteristic curve (AUC) value of 0.958 for the validation set, and a BA of 0.886 and an AUC of 0.947 for the test set. Three external validation sets, including set 1 (3231 FLuc inhibitors and 69 783 noninhibitors), set 2 (695 FLuc inhibitors and 75 913 noninhibitors), and set 3 (1138 FLuc inhibitors and 8155 noninhibitors), were used to verify the predictive ability of our models. The BA values for the three external validation sets given by the best model are 0.864, 0.845, and 0.791, respectively. In addition, the important features or structural fragments related to FLuc inhibitors were recognized by the Shapley additive explanations (SHAP) method along with their influences on predictions, which may provide valuable clues to detecting undesirable luciferase inhibitors. Based on the important and explanatory features, 16 rules were proposed for detecting FLuc inhibitors, which can achieve a correction rate of 70% for FLuc inhibitors. Furthermore, a comparison with existing prediction rules and models for FLuc inhibitors used in virtual screening verified the high reliability of the models and rules proposed in this study. We also used the model to screen three curated chemical databases, and almost 10% of the molecules in the evaluated databases were predicted as inhibitors, highlighting the potential risk of false positives in luciferase-based assays. Finally, a public web server called ChemFLuc was developed (http://admet.scbdd.com/chemfluc/index/), and it offers a free available service to predict potential FLuc inhibitors.
中文翻译:

基于萤光素酶分析的假阳性结果的结构分析和鉴定。
基于萤光素酶的生物发光检测技术在高通量筛选(HTS)中非常受青睐,其中萤火虫萤光素酶(FLuc)是最常用的变体。但是,FLuc抑制剂会干扰荧光素酶的活性,这可能会在HTS分析中导致假阳性信号。为了减少不必要的时间和金钱成本,非常需要用于FLuc抑制剂的计算机模拟模型。在这项研究中,我们建立了由20 888个FLuc抑制剂和198 608个非抑制剂组成的广泛数据集,然后基于三种机器学习(ML)算法和四种分子表示形式的组合,开发了一组分类模型。基于XGBoost和ECFP4以及MOE2d描述符的最佳预测模型得出的平衡精度(BA)为0。验证集为878,接收器工作特性曲线(AUC)值下的面积为0.958,测试集的BA为0.886,AUC为0.947。使用三个外部验证集,包括第1组(3231 FLuc抑制剂和69 783种非抑制剂),第2组(695 FLuc抑制剂和75 913种非抑制剂)和第3组(1138 FLuc抑制剂和8155种非抑制剂)来验证预测的能力。我们的模型。最佳模型给出的三个外部验证集的BA值分别为0.864、0.845和0.791。此外,Shapley加性解释(SHAP)方法已经认识到与FLuc抑制剂相关的重要特征或结构片段,以及它们对预测的影响,这可能为检测不良荧光素酶抑制剂提供有价值的线索。基于重要的解释性特征,提出了检测FLuc抑制剂的16条规则,对FLuc抑制剂的校正率可达到70%。此外,与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。FLuc抑制剂的校正率可达到70%。此外,与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。FLuc抑制剂的校正率可达到70%。此外,与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。强调了在基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。强调了在基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。
更新日期:2020-03-23
中文翻译:

基于萤光素酶分析的假阳性结果的结构分析和鉴定。
基于萤光素酶的生物发光检测技术在高通量筛选(HTS)中非常受青睐,其中萤火虫萤光素酶(FLuc)是最常用的变体。但是,FLuc抑制剂会干扰荧光素酶的活性,这可能会在HTS分析中导致假阳性信号。为了减少不必要的时间和金钱成本,非常需要用于FLuc抑制剂的计算机模拟模型。在这项研究中,我们建立了由20 888个FLuc抑制剂和198 608个非抑制剂组成的广泛数据集,然后基于三种机器学习(ML)算法和四种分子表示形式的组合,开发了一组分类模型。基于XGBoost和ECFP4以及MOE2d描述符的最佳预测模型得出的平衡精度(BA)为0。验证集为878,接收器工作特性曲线(AUC)值下的面积为0.958,测试集的BA为0.886,AUC为0.947。使用三个外部验证集,包括第1组(3231 FLuc抑制剂和69 783种非抑制剂),第2组(695 FLuc抑制剂和75 913种非抑制剂)和第3组(1138 FLuc抑制剂和8155种非抑制剂)来验证预测的能力。我们的模型。最佳模型给出的三个外部验证集的BA值分别为0.864、0.845和0.791。此外,Shapley加性解释(SHAP)方法已经认识到与FLuc抑制剂相关的重要特征或结构片段,以及它们对预测的影响,这可能为检测不良荧光素酶抑制剂提供有价值的线索。基于重要的解释性特征,提出了检测FLuc抑制剂的16条规则,对FLuc抑制剂的校正率可达到70%。此外,与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。FLuc抑制剂的校正率可达到70%。此外,与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。FLuc抑制剂的校正率可达到70%。此外,与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。与用于虚拟筛选的FLuc抑制剂的现有预测规则和模型进行比较,验证了本研究中提出的模型和规则的高度可靠性。我们还使用该模型筛选了三个精选的化学数据库,并且预测评估数据库中几乎10%的分子为抑制剂,从而突出了基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。强调了在基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。强调了在基于荧光素酶的测定中假阳性的潜在风险。最后,开发了一个名为ChemFLuc的公共Web服务器(http://admet.scbdd.com/chemfluc/index/),它提供了免费的可用服务来预测潜在的FLuc抑制剂。











































 京公网安备 11010802027423号
京公网安备 11010802027423号