当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Sequence-specific Quantitation of Mutagenic DNA damage via Polymerase Amplification with an Artificial Nucleotide
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2020-03-20 , DOI: 10.1021/jacs.9b11746 Claudia M N Aloisi 1 , Arman Nilforoushan 1 , Nathalie Ziegler 1 , Shana J Sturla 1
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2020-03-20 , DOI: 10.1021/jacs.9b11746 Claudia M N Aloisi 1 , Arman Nilforoushan 1 , Nathalie Ziegler 1 , Shana J Sturla 1
Affiliation
|
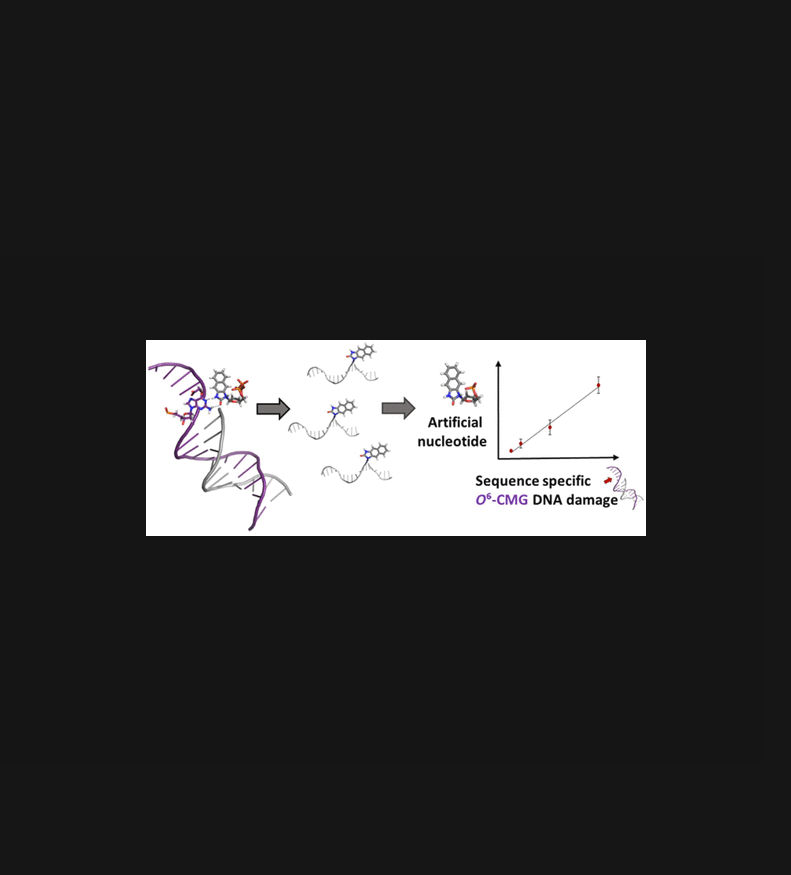
DNA mutations can result from replication errors due to different forms of DNA damage, including low-abundance DNA adducts induced by reactions with electrophiles. The lack of strategies to measure DNA adducts within genomic loci, however, limits our understanding of chemical mutagenesis. The use of artificial nucleotides incorporated opposite DNA adducts by engineered DNA polymerases offers a potential basis for site-specific detection of DNA adducts, but the availability of effective artificial nucleotides that insert opposite DNA adducts is extremely limited, and furthermore, there has been no report of a quantitative strategy for determining how much DNA alkylation occurs in a sequence of interest. In this work, we synthesized an artificial nucleotide triphosphate that is selectively inserted opposite O6-carboxymethyl-guanine DNA by an engineered polymerase and is required for DNA synthesis past the adduct. We characterized the mechanism of this enzymatic process and demonstrated that the artificial nucleotide is a marker for the presence and location in the genome of O6-carboxymethyl-guanine. Finally, we established a mass spectrometric method for quantifying the incorporated artificial nucleotide and obtained a linear relationship with the amount of O6-carboxymethyl-guanine in the target sequence. In this work, we present a strategy to identify, locate, and quantify a mutagenic DNA adduct, advancing tools for linking DNA alkylation to mutagenesis and for detecting DNA adducts in genes as potential diagnostic biomarkers for cancer prevention.
中文翻译:

用人工核苷酸通过聚合酶扩增对诱变 DNA 损伤进行序列特异性定量
DNA 突变可能是由于不同形式的 DNA 损伤引起的复制错误造成的,包括与亲电试剂反应引起的低丰度 DNA 加合物。然而,缺乏测量基因组位点内 DNA 加合物的策略限制了我们对化学诱变的理解。使用通过工程 DNA 聚合酶掺入反向 DNA 加合物的人工核苷酸为 DNA 加合物的位点特异性检测提供了潜在基础,但插入反向 DNA 加合物的有效人工核苷酸的可用性极其有限,此外,还没有报道用于确定感兴趣序列中发生多少 DNA 烷基化的定量策略。在这项工作中,我们合成了一种人工三磷酸核苷酸,它通过工程聚合酶选择性地插入到 O6-羧甲基-鸟嘌呤 DNA 的对面,并且是 DNA 合成通过加合物所必需的。我们表征了这种酶促过程的机制,并证明人工核苷酸是 O6-羧甲基-鸟嘌呤基因组中存在和位置的标记。最后,我们建立了一种用于量化掺入的人工核苷酸的质谱方法,并获得了与目标序列中 O6-羧甲基-鸟嘌呤含量的线性关系。在这项工作中,我们提出了一种识别、定位和量化诱变 DNA 加合物的策略,改进了将 DNA 烷基化与诱变联系起来的工具,以及检测基因中的 DNA 加合物作为预防癌症的潜在诊断生物标志物的工具。
更新日期:2020-03-20
中文翻译:

用人工核苷酸通过聚合酶扩增对诱变 DNA 损伤进行序列特异性定量
DNA 突变可能是由于不同形式的 DNA 损伤引起的复制错误造成的,包括与亲电试剂反应引起的低丰度 DNA 加合物。然而,缺乏测量基因组位点内 DNA 加合物的策略限制了我们对化学诱变的理解。使用通过工程 DNA 聚合酶掺入反向 DNA 加合物的人工核苷酸为 DNA 加合物的位点特异性检测提供了潜在基础,但插入反向 DNA 加合物的有效人工核苷酸的可用性极其有限,此外,还没有报道用于确定感兴趣序列中发生多少 DNA 烷基化的定量策略。在这项工作中,我们合成了一种人工三磷酸核苷酸,它通过工程聚合酶选择性地插入到 O6-羧甲基-鸟嘌呤 DNA 的对面,并且是 DNA 合成通过加合物所必需的。我们表征了这种酶促过程的机制,并证明人工核苷酸是 O6-羧甲基-鸟嘌呤基因组中存在和位置的标记。最后,我们建立了一种用于量化掺入的人工核苷酸的质谱方法,并获得了与目标序列中 O6-羧甲基-鸟嘌呤含量的线性关系。在这项工作中,我们提出了一种识别、定位和量化诱变 DNA 加合物的策略,改进了将 DNA 烷基化与诱变联系起来的工具,以及检测基因中的 DNA 加合物作为预防癌症的潜在诊断生物标志物的工具。











































 京公网安备 11010802027423号
京公网安备 11010802027423号