当前位置:
X-MOL 学术
›
Syst. Entomol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Phylogenomic analysis of the beetle suborder Adephaga with comparison of tailored and generalized ultraconserved element probe performance
Systematic Entomology ( IF 4.7 ) Pub Date : 2019-12-19 , DOI: 10.1111/syen.12413 Grey T. Gustafson 1, 2 , Stephen M. Baca 1, 2 , Alana M. Alexander 3 , Andrew E. Z. Short 1, 2
Systematic Entomology ( IF 4.7 ) Pub Date : 2019-12-19 , DOI: 10.1111/syen.12413 Grey T. Gustafson 1, 2 , Stephen M. Baca 1, 2 , Alana M. Alexander 3 , Andrew E. Z. Short 1, 2
Affiliation
|
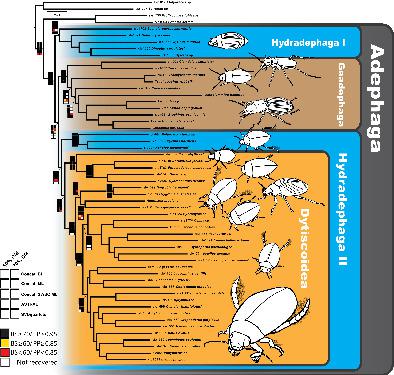
Adephaga is the second largest suborder of beetles (Coleoptera) and they serve as important arthropod predators in both aquatic and terrestrial ecosystems. The suborder is divided into Geadephaga comprising terrestrial families and Hydradephaga for aquatic lineages. Despite numerous studies, phylogenetic relationships among the adephagan families and monophyly of the Hydradephaga itself remain in question. Here we conduct a comprehensive phylogenomic analysis of the suborder using ultraconserved elements (UCEs). This study presents the first in vitro test of a newly developed UCE probe set customized for use within Adephaga that includes both probes tailored specifically for the suborder, alongside generalized Coleoptera probes previously found to work in adephagan taxa. We assess the utility of the entire probe set, as well as comparing the tailored and generalized probes alone for reconstructing evolutionary relationships. Our analyses recovered strong support for the paraphyly of Hydradephaga with whirligig beetles (Gyrinidae) placed as sister to all other adephagan families. Geadephaga was strongly supported as monophyletic and placed sister to a clade composed of Haliplidae + Dytiscoidea. Monophyly of Dytiscoidea was strongly supported with relationships among the dytiscoid families resolved and strongly supported. Relationships among the subfamilies of Dytiscidae were strongly supported but largely incongruent with prior phylogenetic estimates for the family. The results of our UCE probe comparison showed that tailored probes alone outperformed generalized probes alone, as well as the full combined probe set (containing both types of probes), under decreased taxon sampling. When taxon sampling was increased, the full combined probe set outperformed both tailored probes and generalized probes alone. This study provides further evidence that UCE probe sets customized for a focal group result in a greater number of recovered loci and substantially improve phylogenomic analysis.
中文翻译:

甲虫亚目 Adephaga 的系统发育分析与定制和广义超保守元素探针性能的比较
Adephaga 是甲虫(鞘翅目)的第二大亚目,它们是水生和陆地生态系统中重要的节肢动物捕食者。该亚目分为包括陆生科的 Geadephaga 和用于水生谱系的 Hydradephaga。尽管进行了大量研究,但 adephagan 家族之间的系统发育关系和 Hydradephaga 本身的单一系统仍然存在问题。在这里,我们使用超保守元素 (UCE) 对亚目进行了全面的系统发育分析。这项研究展示了新开发的 UCE 探针组的首次体外测试,该探针组定制用于 Adephaga,其中包括专为该亚目定制的两种探针,以及先前发现在 adephagan 分类群中工作的广义鞘翅目探针。我们评估整个探针组的效用,以及单独比较定制和广义探针以重建进化关系。我们的分析恢复了对 Hydradephaga 副系的强有力支持,其中旋转甲虫(Gyrinidae)被放置为所有其他 adephagan 家族的姐妹。Geadephaga 被强烈支持为单系的,并且是由 Haliplidae + Dytiscoidea 组成的进化枝的姐妹。Dytiscoidea 的单系得到了强烈支持,并且解决并强烈支持了 dytiscoidea 科之间的关系。龙牙科亚科之间的关系得到强烈支持,但在很大程度上与先前对该科的系统发育估计不一致。我们的 UCE 探针比较的结果表明,单独的定制探针优于单独的广义探针,以及完整的组合探针组(包含两种类型的探针),在减少的分类单元抽样下。当分类单元采样增加时,完整的组合探针集优于单独的定制探针和广义探针。这项研究提供了进一步的证据,表明为焦点组定制的 UCE 探针集会导致更多的恢复基因座并显着改善系统发育分析。
更新日期:2019-12-19
中文翻译:

甲虫亚目 Adephaga 的系统发育分析与定制和广义超保守元素探针性能的比较
Adephaga 是甲虫(鞘翅目)的第二大亚目,它们是水生和陆地生态系统中重要的节肢动物捕食者。该亚目分为包括陆生科的 Geadephaga 和用于水生谱系的 Hydradephaga。尽管进行了大量研究,但 adephagan 家族之间的系统发育关系和 Hydradephaga 本身的单一系统仍然存在问题。在这里,我们使用超保守元素 (UCE) 对亚目进行了全面的系统发育分析。这项研究展示了新开发的 UCE 探针组的首次体外测试,该探针组定制用于 Adephaga,其中包括专为该亚目定制的两种探针,以及先前发现在 adephagan 分类群中工作的广义鞘翅目探针。我们评估整个探针组的效用,以及单独比较定制和广义探针以重建进化关系。我们的分析恢复了对 Hydradephaga 副系的强有力支持,其中旋转甲虫(Gyrinidae)被放置为所有其他 adephagan 家族的姐妹。Geadephaga 被强烈支持为单系的,并且是由 Haliplidae + Dytiscoidea 组成的进化枝的姐妹。Dytiscoidea 的单系得到了强烈支持,并且解决并强烈支持了 dytiscoidea 科之间的关系。龙牙科亚科之间的关系得到强烈支持,但在很大程度上与先前对该科的系统发育估计不一致。我们的 UCE 探针比较的结果表明,单独的定制探针优于单独的广义探针,以及完整的组合探针组(包含两种类型的探针),在减少的分类单元抽样下。当分类单元采样增加时,完整的组合探针集优于单独的定制探针和广义探针。这项研究提供了进一步的证据,表明为焦点组定制的 UCE 探针集会导致更多的恢复基因座并显着改善系统发育分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号