当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Reconciling NMR Structures of the HIV-1 Nucleocapsid Protein NCp7 Using Extensive Polarizable Force Field Free-Energy Simulations.
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2020-03-16 , DOI: 10.1021/acs.jctc.9b01204 Léa El Khoury 1, 2 , Frédéric Célerse 1, 3 , Louis Lagardère 4, 5 , Luc-Henri Jolly 4 , Etienne Derat 3 , Zeina Hobaika 2 , Richard G Maroun 2 , Pengyu Ren 6 , Serge Bouaziz 7 , Nohad Gresh 1 , Jean-Philip Piquemal 1, 6, 8
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2020-03-16 , DOI: 10.1021/acs.jctc.9b01204 Léa El Khoury 1, 2 , Frédéric Célerse 1, 3 , Louis Lagardère 4, 5 , Luc-Henri Jolly 4 , Etienne Derat 3 , Zeina Hobaika 2 , Richard G Maroun 2 , Pengyu Ren 6 , Serge Bouaziz 7 , Nohad Gresh 1 , Jean-Philip Piquemal 1, 6, 8
Affiliation
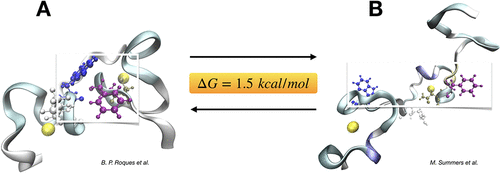
|
Using polarizable (AMOEBA) and nonpolarizable (CHARMM) force fields, we compare the relative free energy stability of two extreme conformations of the HIV-1 nucleocapsid protein NCp7 that had been previously experimentally advocated to prevail in solution. Using accelerated sampling techniques, we show that they differ in stability by no more than 0.75–1.9 kcal/mol depending on the reference protein sequence. While the extended form appears to be the most probable structure, both forms should thus coexist in water explaining the differing NMR findings.
中文翻译:

使用广泛的极化力场自由能模拟,对HIV-1核蛋白蛋白NCp7的NMR结构进行核对。
使用极化的(AMOEBA)和非极化的(CHARMM)力场,我们比较了HIV-1核衣壳蛋白NCp7的两个极端构象的相对自由能稳定性,该构象先前曾通过实验提倡在溶液中盛行。使用加速采样技术,我们发现它们的稳定性差异不超过0.75–1.9 kcal / mol,具体取决于参考蛋白质序列。虽然扩展形式似乎是最可能的结构,但两种形式都应共存于水中,从而解释了不同的NMR发现。
更新日期:2020-04-24
中文翻译:

使用广泛的极化力场自由能模拟,对HIV-1核蛋白蛋白NCp7的NMR结构进行核对。
使用极化的(AMOEBA)和非极化的(CHARMM)力场,我们比较了HIV-1核衣壳蛋白NCp7的两个极端构象的相对自由能稳定性,该构象先前曾通过实验提倡在溶液中盛行。使用加速采样技术,我们发现它们的稳定性差异不超过0.75–1.9 kcal / mol,具体取决于参考蛋白质序列。虽然扩展形式似乎是最可能的结构,但两种形式都应共存于水中,从而解释了不同的NMR发现。



























 京公网安备 11010802027423号
京公网安备 11010802027423号