当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Molecular Dynamics Simulations Based on Newly Developed Force Field Parameters for Cu2+ Spin Labels Provide Insights into Double-Histidine-Based Double Electron–Electron Resonance
The Journal of Physical Chemistry B ( IF 3.3 ) Pub Date : 2020-03-26 , DOI: 10.1021/acs.jpcb.0c00739 Xiaowei Bogetti 1 , Shreya Ghosh 1 , Austin Gamble Jarvi 1 , Junmei Wang 2 , Sunil Saxena 1
The Journal of Physical Chemistry B ( IF 3.3 ) Pub Date : 2020-03-26 , DOI: 10.1021/acs.jpcb.0c00739 Xiaowei Bogetti 1 , Shreya Ghosh 1 , Austin Gamble Jarvi 1 , Junmei Wang 2 , Sunil Saxena 1
Affiliation
|
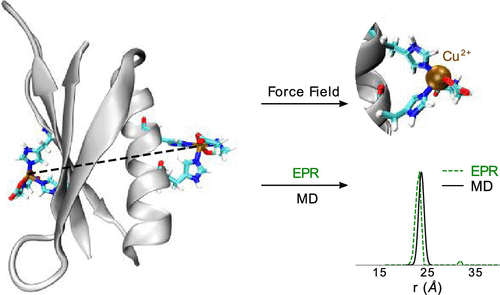
Electron paramagnetic resonance (EPR) in combination with the recently developed double-histidine (dHis)-based Cu2+ spin labeling has provided valuable insights into protein structure and conformational dynamics. To relate sparse distance constraints measured by EPR to protein fluctuations in solution, modeling techniques are needed. In this work, we have developed force field parameters for Cu2+–nitrilotriacetic and Cu2+–iminodiacetic acid spin labels. We employed molecular dynamics (MD) simulations to capture the atomic-level details of dHis-labeled protein fluctuations. The interspin distances extracted from 200 ns MD trajectories show good agreement with the experimental results. The MD simulations also illustrate the dramatic rigidity of the Cu2+ labels compared to the standard nitroxide spin label. Further, the relative orientations between spin-labeled sites were measured to provide insight into the use of double electron–electron resonance (DEER) methods for such labels. The relative mean angles, as well as the standard deviations of the relative angles, agree well in general with the spectral simulations published previously. The fluctuations of relative orientations help rationalize why orientation selectivity effects are minimal at X-band frequencies, but observable at the Q-band for such labels. In summary, the results show that by combining the experimental results with MD simulations precise information about protein conformations as well as flexibility can be obtained.
中文翻译:

基于新开发的Cu 2+自旋标记力场参数的分子动力学模拟为基于双组氨酸的双电子-电子共振提供了见识
电子顺磁共振(EPR)与最近开发的基于双组氨酸(dHis)的Cu 2+自旋标记相结合,提供了对蛋白质结构和构象动力学的有价值的见解。为了将通过EPR测量的稀疏距离约束与溶液中的蛋白质波动相关联,需要建模技术。在这项工作中,我们对铜发达力场参数2+次氮基三和Cu 2+亚氨基二乙酸自旋标记。我们采用分子动力学(MD)模拟来捕获dHis标记的蛋白质波动的原子级细节。从200 ns MD轨迹提取的自旋间距离与实验结果显示出良好的一致性。MD模拟还说明了Cu的惊人刚性与标准的一氧化氮旋转标签相比,具有2+个标签。此外,对自旋标记位点之间的相对方向进行了测量,以深入了解使用双电子电子共振(DEER)方法进行此类标记。相对平均角度以及相对角度的标准偏差通常与先前发布的光谱模拟完全吻合。相对方向的波动有助于合理化为什么为什么方向选择性效应在X波段频率上最小,而在Q波段这种标签可见。总之,结果表明,通过将实验结果与MD模拟相结合,可以获得有关蛋白质构象以及柔韧性的精确信息。
更新日期:2020-03-27
中文翻译:

基于新开发的Cu 2+自旋标记力场参数的分子动力学模拟为基于双组氨酸的双电子-电子共振提供了见识
电子顺磁共振(EPR)与最近开发的基于双组氨酸(dHis)的Cu 2+自旋标记相结合,提供了对蛋白质结构和构象动力学的有价值的见解。为了将通过EPR测量的稀疏距离约束与溶液中的蛋白质波动相关联,需要建模技术。在这项工作中,我们对铜发达力场参数2+次氮基三和Cu 2+亚氨基二乙酸自旋标记。我们采用分子动力学(MD)模拟来捕获dHis标记的蛋白质波动的原子级细节。从200 ns MD轨迹提取的自旋间距离与实验结果显示出良好的一致性。MD模拟还说明了Cu的惊人刚性与标准的一氧化氮旋转标签相比,具有2+个标签。此外,对自旋标记位点之间的相对方向进行了测量,以深入了解使用双电子电子共振(DEER)方法进行此类标记。相对平均角度以及相对角度的标准偏差通常与先前发布的光谱模拟完全吻合。相对方向的波动有助于合理化为什么为什么方向选择性效应在X波段频率上最小,而在Q波段这种标签可见。总之,结果表明,通过将实验结果与MD模拟相结合,可以获得有关蛋白质构象以及柔韧性的精确信息。



























 京公网安备 11010802027423号
京公网安备 11010802027423号