当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Accurate Identification of Deamidation and Citrullination from Global Shotgun Proteomics Data Using a Dual-Search Delta Score Strategy.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-03-23 , DOI: 10.1021/acs.jproteome.9b00766 Xi Wang 1, 2 , Adam C Swensen 1 , Tong Zhang 1 , Paul D Piehowski 1 , Matthew J Gaffrey 1 , Matthew E Monroe 1 , Ying Zhu 3 , Hailiang Dong 2 , Wei-Jun Qian 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-03-23 , DOI: 10.1021/acs.jproteome.9b00766 Xi Wang 1, 2 , Adam C Swensen 1 , Tong Zhang 1 , Paul D Piehowski 1 , Matthew J Gaffrey 1 , Matthew E Monroe 1 , Ying Zhu 3 , Hailiang Dong 2 , Wei-Jun Qian 1
Affiliation
|
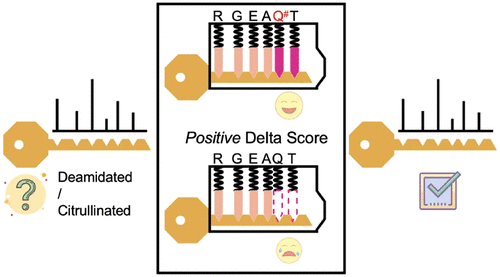
Proteins with deamidated/citrullinated amino acids play critical roles in the pathogenesis of many human diseases; however, identifying these modifications in complex biological samples has been an ongoing challenge. Herein we present a method to accurately identify these modifications from shotgun proteomics data generated by a deep proteome profiling study of human pancreatic islets obtained by laser capture microdissection. All MS/MS spectra were searched twice using MSGF+ database matching, with and without a dynamic +0.9840 Da mass shift modification on amino acids asparagine, glutamine, and arginine (NQR). Consequently, each spectrum generates two peptide-to-spectrum matches (PSMs) with MSGF+ scores, which were used for the Delta Score calculation. It was observed that all PSMs with positive Delta Score values were clustered with mass errors around 0 ppm, while PSMs with negative Delta Score values were distributed nearly equally within the defined mass error range (20 ppm) for database searching. To estimate false discovery rate (FDR) of modified peptides, a "target-mock" strategy was applied in which data sets were searched against a concatenated database containing "real-modified" (+0.9840 Da) and "mock-modified" (+1.0227 Da) peptide masses. The FDR was controlled to ∼2% using a Delta Score filter value greater than zero. Manual inspection of spectra showed that PSMs with positive Delta Score values contained deamidated/citrullinated fragments in their MS/MS spectra. Many citrullinated sites identified in this study were biochemically confirmed as autoimmunogenic epitopes of autoimmune diseases in literature. The results demonstrated that in situ deamidated/citrullinated peptides can be accurately identified from shotgun tissue proteomics data using this dual-search Delta Score strategy. Raw MS data is available at ProteomeXchange (PXD010150).
中文翻译:

使用双重搜索Delta评分策略从全球散弹枪蛋白质组学数据中准确识别脱酰胺和瓜耳化。
具有酰胺化/瓜氨酸化氨基酸的蛋白质在许多人类疾病的发病机理中起着至关重要的作用。然而,鉴定复杂生物样品中的这些修饰一直是一项持续的挑战。本文中,我们提出了一种方法,可以从通过激光捕获显微切割技术获得的人胰岛的深层蛋白质组分析研究生成的shot弹枪蛋白质组学数据中,准确识别这些修饰。使用MSGF +数据库匹配对所有MS / MS谱图进行了两次搜索,对天冬酰胺,谷氨酰胺和精氨酸(NQR)进行动态和不进行动态+0.9840 Da质量转移修饰。因此,每个光谱都会生成两个具有MSGF +分数的肽谱匹配(PSM),用于Delta分数计算。可以看到,所有具有正Delta得分值的PSM都聚集有质量误差大约为0 ppm的聚类,而具有负Delta得分值的PSM几乎平均地分布在定义的质量误差范围内(20 ppm),以进行数据库搜索。为了估算修饰肽的错误发现率(FDR),应用了“靶点模拟”策略,其中针对包含“真实修饰的”(+0.9840 Da)和“模拟修饰的”(+ 1.0227 Da)肽质量。使用大于零的Delta Score过滤器值将FDR控制为〜2%。手动检查光谱表明,具有正Delta得分值的PSM在其MS / MS光谱中包含脱酰胺/瓜氨酸化的碎片。在本研究中鉴定出的许多瓜氨酸化位点在生化方面被文献确定为自身免疫性疾病的自身免疫原性表位。结果表明,使用这种双重搜索Delta评分策略,可以从shot弹枪组织蛋白质组学数据中准确识别原位脱酰胺基/瓜氨酸化肽。原始MS数据可从ProteomeXchange(PXD010150)获得。
更新日期:2020-04-24
中文翻译:

使用双重搜索Delta评分策略从全球散弹枪蛋白质组学数据中准确识别脱酰胺和瓜耳化。
具有酰胺化/瓜氨酸化氨基酸的蛋白质在许多人类疾病的发病机理中起着至关重要的作用。然而,鉴定复杂生物样品中的这些修饰一直是一项持续的挑战。本文中,我们提出了一种方法,可以从通过激光捕获显微切割技术获得的人胰岛的深层蛋白质组分析研究生成的shot弹枪蛋白质组学数据中,准确识别这些修饰。使用MSGF +数据库匹配对所有MS / MS谱图进行了两次搜索,对天冬酰胺,谷氨酰胺和精氨酸(NQR)进行动态和不进行动态+0.9840 Da质量转移修饰。因此,每个光谱都会生成两个具有MSGF +分数的肽谱匹配(PSM),用于Delta分数计算。可以看到,所有具有正Delta得分值的PSM都聚集有质量误差大约为0 ppm的聚类,而具有负Delta得分值的PSM几乎平均地分布在定义的质量误差范围内(20 ppm),以进行数据库搜索。为了估算修饰肽的错误发现率(FDR),应用了“靶点模拟”策略,其中针对包含“真实修饰的”(+0.9840 Da)和“模拟修饰的”(+ 1.0227 Da)肽质量。使用大于零的Delta Score过滤器值将FDR控制为〜2%。手动检查光谱表明,具有正Delta得分值的PSM在其MS / MS光谱中包含脱酰胺/瓜氨酸化的碎片。在本研究中鉴定出的许多瓜氨酸化位点在生化方面被文献确定为自身免疫性疾病的自身免疫原性表位。结果表明,使用这种双重搜索Delta评分策略,可以从shot弹枪组织蛋白质组学数据中准确识别原位脱酰胺基/瓜氨酸化肽。原始MS数据可从ProteomeXchange(PXD010150)获得。







































 京公网安备 11010802027423号
京公网安备 11010802027423号