当前位置:
X-MOL 学术
›
Commun. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Deep splicing plasticity of the human adenovirus type 5 transcriptome drives virus evolution.
Communications Biology ( IF 5.2 ) Pub Date : 2020-03-13 , DOI: 10.1038/s42003-020-0849-9 I'ah Donovan-Banfield 1 , Andrew S Turnell 2 , Julian A Hiscox 3 , Keith N Leppard 4 , David A Matthews 1
Communications Biology ( IF 5.2 ) Pub Date : 2020-03-13 , DOI: 10.1038/s42003-020-0849-9 I'ah Donovan-Banfield 1 , Andrew S Turnell 2 , Julian A Hiscox 3 , Keith N Leppard 4 , David A Matthews 1
Affiliation
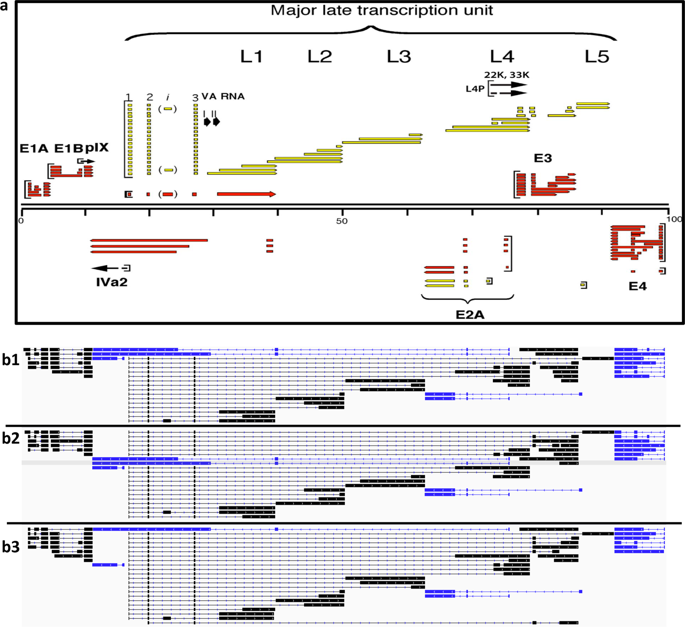
|
Viral genomes have high gene densities and complex transcription strategies rendering transcriptome analysis through short-read RNA-seq approaches problematic. Adenovirus transcription and splicing is especially complex. We used long-read direct RNA sequencing to study adenovirus transcription and splicing during infection. This revealed a previously unappreciated complexity of alternative splicing and potential for secondary initiating codon usage. Moreover, we find that most viral transcripts tend to shorten polyadenylation lengths as infection progresses. Development of an open reading frame centric bioinformatics analysis pipeline provided a deeper quantitative and qualitative understanding of adenovirus's genetic potential. Across the viral genome adenovirus makes multiple distinctly spliced transcripts that code for the same protein. Over 11,000 different splicing patterns were recorded across the viral genome, most occurring at low levels. This low-level use of alternative splicing patterns potentially enables the virus to maximise its coding potential over evolutionary timescales.
中文翻译:

人类 5 型腺病毒转录组的深度剪接可塑性驱动病毒进化。
病毒基因组具有高基因密度和复杂的转录策略,使得通过短读长 RNA-seq 方法进行转录组分析存在问题。腺病毒转录和剪接尤其复杂。我们使用长读长直接 RNA 测序来研究感染期间的腺病毒转录和剪接。这揭示了先前未被认识到的选择性剪接的复杂性以及二次起始密码子使用的潜力。此外,我们发现随着感染的进展,大多数病毒转录本倾向于缩短多腺苷酸化长度。以开放阅读框为中心的生物信息学分析流程的开发提供了对腺病毒遗传潜力的更深入的定量和定性理解。腺病毒在病毒基因组中产生多个明显剪接的转录本,编码相同的蛋白质。整个病毒基因组记录了超过 11,000 种不同的剪接模式,大多数发生在低水平。这种选择性剪接模式的低水平使用可能使病毒在进化时间尺度上最大化其编码潜力。
更新日期:2020-03-16
中文翻译:

人类 5 型腺病毒转录组的深度剪接可塑性驱动病毒进化。
病毒基因组具有高基因密度和复杂的转录策略,使得通过短读长 RNA-seq 方法进行转录组分析存在问题。腺病毒转录和剪接尤其复杂。我们使用长读长直接 RNA 测序来研究感染期间的腺病毒转录和剪接。这揭示了先前未被认识到的选择性剪接的复杂性以及二次起始密码子使用的潜力。此外,我们发现随着感染的进展,大多数病毒转录本倾向于缩短多腺苷酸化长度。以开放阅读框为中心的生物信息学分析流程的开发提供了对腺病毒遗传潜力的更深入的定量和定性理解。腺病毒在病毒基因组中产生多个明显剪接的转录本,编码相同的蛋白质。整个病毒基因组记录了超过 11,000 种不同的剪接模式,大多数发生在低水平。这种选择性剪接模式的低水平使用可能使病毒在进化时间尺度上最大化其编码潜力。









































 京公网安备 11010802027423号
京公网安备 11010802027423号