当前位置:
X-MOL 学术
›
J. Biomed. Inform.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Empirically-derived synthetic populations to mitigate small sample sizes.
Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2020-03-12 , DOI: 10.1016/j.jbi.2020.103408 Erin E Fowler 1 , Anders Berglund 2 , Michael J Schell 2 , Thomas A Sellers , Steven Eschrich 2 , John Heine 1
Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2020-03-12 , DOI: 10.1016/j.jbi.2020.103408 Erin E Fowler 1 , Anders Berglund 2 , Michael J Schell 2 , Thomas A Sellers , Steven Eschrich 2 , John Heine 1
Affiliation
|
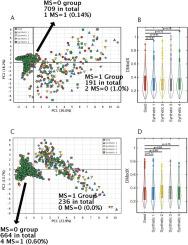
Limited sample sizes can lead to spurious modeling findings in biomedical research. The objective of this work is to present a new method to generate synthetic populations (SPs) from limited samples using matched case-control data (n = 180 pairs), considered as two separate limited samples. SPs were generated with multivariate kernel density estimations (KDEs) with unconstrained bandwidth matrices. We included four continuous variables and one categorical variable for each individual. Bandwidth matrices were determined with Differential Evolution (DE) optimization by covariance comparisons. Four synthetic samples (n = 180) were derived from their respective SPs. Similarity between observed samples with synthetic samples was compared assuming their empirical probability density functions (EPDFs) were similar. EPDFs were compared with the maximum mean discrepancy (MMD) test statistic based on the Kernel Two-Sample Test. To evaluate similarity within a modeling context, EPDFs derived from the Principal Component Analysis (PCA) scores and residuals were summarized with the distance to the model in X-space (DModX) as additional comparisons. Four SPs were generated from each sample. The probability of selecting a replicate when randomly constructing synthetic samples (n = 180) was infinitesimally small. MMD tests indicated that the observed sample EPDFs were similar to the respective synthetic EPDFs. For the samples, PCA scores and residuals did not deviate significantly when compared with their respective synthetic samples. The feasibility of this approach was demonstrated by producing synthetic data at the individual level, statistically similar to the observed samples. The methodology coupled KDE with DE optimization and deployed novel similarity metrics derived from PCA. This approach could be used to generate larger-sized synthetic samples. To develop this approach into a research tool for data exploration purposes, additional evaluation with increased dimensionality is required. Moreover, given a fully specified population, the degree to which individuals can be discarded while synthesizing the respective population accurately will be investigated. When these objectives are addressed, comparisons with other techniques such as bootstrapping will be required for a complete evaluation.
中文翻译:

以经验为依据的合成种群可减少小样本量。
有限的样本量可能导致生物医学研究中虚假的建模发现。这项工作的目的是提出一种新方法,使用匹配的病例对照数据(n = 180对)从有限的样本中生成合成种群(SP),该数据被视为两个单独的有限样本。SP是使用具有不受约束的带宽矩阵的多变量内核密度估计(KDE)生成的。我们为每个人包括了四个连续变量和一个分类变量。带宽矩阵通过协方差比较通过差分演化(DE)优化来确定。四个合成样本(n = 180)分别来自其SP。假设它们的经验概率密度函数(EPDF)相似,则比较观察到的样品与合成样品之间的相似性。将EPDF与基于内核两次抽样检验的最大平均差异(MMD)检验统计数据进行比较。为了评估建模上下文中的相似性,总结了从主成分分析(PCA)分数和残差得出的EPDF,并在X空间(DModX)中与模型的距离进行了比较。从每个样品产生四个SP。随机构建合成样本(n = 180)时选择重复样本的可能性极小。MMD测试表明,观察到的样品EPDF与各自的合成EPDF相似。对于样品,与它们各自的合成样品相比,PCA分数和残差没有显着偏离。通过在各个级别生成综合数据,证明了该方法的可行性,统计上与观察到的样本相似。该方法将KDE与DE优化结合在一起,并部署了从PCA派生的新颖相似性指标。该方法可用于生成较大尺寸的合成样品。为了将此方法发展为用于数据探索目的的研究工具,需要进行附加评估,并增加维度。而且,在给定完全指定的种群的情况下,将研究在准确合成各个种群的同时可以丢弃个体的程度。解决这些目标时,将需要与其他技术(例如自举)进行比较,以进行全面评估。该方法可用于生成较大尺寸的合成样品。为了将此方法发展为用于数据探索目的的研究工具,需要进行附加评估,并增加维度。而且,在给定完全指定的种群的情况下,将研究在准确合成各个种群的同时可以丢弃个体的程度。解决了这些目标后,将需要与其他技术(例如自举)进行比较,以进行全面评估。该方法可用于生成较大尺寸的合成样品。为了将此方法发展为用于数据探索目的的研究工具,需要进行附加评估,并增加维度。而且,在给定完全指定的种群的情况下,将研究在准确合成各个种群的同时可以丢弃个体的程度。解决了这些目标后,将需要与其他技术(例如自举)进行比较,以进行全面评估。
更新日期:2020-04-21
中文翻译:

以经验为依据的合成种群可减少小样本量。
有限的样本量可能导致生物医学研究中虚假的建模发现。这项工作的目的是提出一种新方法,使用匹配的病例对照数据(n = 180对)从有限的样本中生成合成种群(SP),该数据被视为两个单独的有限样本。SP是使用具有不受约束的带宽矩阵的多变量内核密度估计(KDE)生成的。我们为每个人包括了四个连续变量和一个分类变量。带宽矩阵通过协方差比较通过差分演化(DE)优化来确定。四个合成样本(n = 180)分别来自其SP。假设它们的经验概率密度函数(EPDF)相似,则比较观察到的样品与合成样品之间的相似性。将EPDF与基于内核两次抽样检验的最大平均差异(MMD)检验统计数据进行比较。为了评估建模上下文中的相似性,总结了从主成分分析(PCA)分数和残差得出的EPDF,并在X空间(DModX)中与模型的距离进行了比较。从每个样品产生四个SP。随机构建合成样本(n = 180)时选择重复样本的可能性极小。MMD测试表明,观察到的样品EPDF与各自的合成EPDF相似。对于样品,与它们各自的合成样品相比,PCA分数和残差没有显着偏离。通过在各个级别生成综合数据,证明了该方法的可行性,统计上与观察到的样本相似。该方法将KDE与DE优化结合在一起,并部署了从PCA派生的新颖相似性指标。该方法可用于生成较大尺寸的合成样品。为了将此方法发展为用于数据探索目的的研究工具,需要进行附加评估,并增加维度。而且,在给定完全指定的种群的情况下,将研究在准确合成各个种群的同时可以丢弃个体的程度。解决这些目标时,将需要与其他技术(例如自举)进行比较,以进行全面评估。该方法可用于生成较大尺寸的合成样品。为了将此方法发展为用于数据探索目的的研究工具,需要进行附加评估,并增加维度。而且,在给定完全指定的种群的情况下,将研究在准确合成各个种群的同时可以丢弃个体的程度。解决了这些目标后,将需要与其他技术(例如自举)进行比较,以进行全面评估。该方法可用于生成较大尺寸的合成样品。为了将此方法发展为用于数据探索目的的研究工具,需要进行附加评估,并增加维度。而且,在给定完全指定的种群的情况下,将研究在准确合成各个种群的同时可以丢弃个体的程度。解决了这些目标后,将需要与其他技术(例如自举)进行比较,以进行全面评估。











































 京公网安备 11010802027423号
京公网安备 11010802027423号