当前位置:
X-MOL 学术
›
Environ. Sci.: Processes Impacts
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Insights into origins and function of the unexplored majority of the reductive dehalogenase gene family as a result of genome assembly and ortholog group classification.
Environmental Science: Processes & Impacts ( IF 4.3 ) Pub Date : 2020-03-11 , DOI: 10.1039/c9em00605b Olivia Molenda 1 , Luz A Puentes Jácome , Xuan Cao , Camilla L Nesbø , Shuiquan Tang , Nadia Morson , Jonas Patron , Line Lomheim , David S Wishart , Elizabeth A Edwards
Environmental Science: Processes & Impacts ( IF 4.3 ) Pub Date : 2020-03-11 , DOI: 10.1039/c9em00605b Olivia Molenda 1 , Luz A Puentes Jácome , Xuan Cao , Camilla L Nesbø , Shuiquan Tang , Nadia Morson , Jonas Patron , Line Lomheim , David S Wishart , Elizabeth A Edwards
Affiliation
|
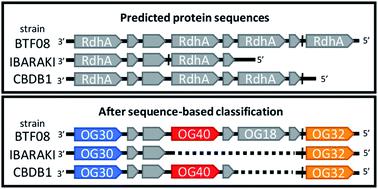
Organohalide respiring bacteria (OHRB) express reductive dehalogenases for energy conservation and growth. Some of these enzymes catalyze the reductive dehalogenation of chlorinated and brominated pollutants in anaerobic subsurface environments, providing a valuable ecosystem service. Dehalococcoides mccartyi strains have been most extensively studied owing to their ability to dechlorinate all chlorinated ethenes – most notably carcinogenic vinyl chloride – to ethene. The genomes of OHRB, particularly obligate OHRB, often harbour multiple putative reductive dehalogenase genes (rdhA), most of which have yet to be characterized. We recently sequenced and closed the genomes of eight new strains, increasing the number of available D. mccartyi genomes in NCBI from 16 to 24. From all available OHRB genomes, we classified predicted translations of reductive dehalogenase genes using a previously established 90% amino acid pairwise identity cut-off to identify Ortholog Groups (OGs). Interestingly, the majority of D. mccartyi dehalogenase gene sequences, once classified into OGs, exhibited a remarkable degree of synteny (gene order) in all genomes sequenced to date. This organization was not apparent without the classification. A high degree of synteny indicates that differences arose from rdhA gene loss rather than recombination. Phylogenetic analysis suggests that most rdhA genes have a long evolutionary history in the Dehalococcoidia with origin prior to speciation of Dehalococcoides and Dehalogenimonas. We also looked for evidence of synteny in the genomes of other species of OHRB. Unfortunately, there are too few closed Dehalogenimonas genomes to compare at this time. There is some partial evidence for synteny in the Dehalobacter restrictus genomes, but here too more closed genomes are needed for confirmation. Interestingly, we found that the rdhA genes that encode enzymes that catalyze dehalogenation of industrial pollutants are the only rdhA genes with strong evidence of recent lateral transfer – at least in the genomes examined herein. Given the utility of the RdhA sequence classification to comparative analyses, we are building a public web server (http://RDaseDB.biozone.utoronto.ca) for the community to use, which allows users to add and classify new sequences, and download the entire curated database of reductive dehalogenases.
中文翻译:

通过基因组组装和直系同源物组分类,了解还原性脱卤化氢酶基因家族的未开发多数的起源和功能。
有机卤化物呼吸细菌(OHRB)表达还原性脱卤素酶,用于能量保存和生长。这些酶中的某些可在厌氧地下环境中催化氯化物和溴化污染物的还原脱卤作用,从而提供有价值的生态系统服务。麦卡迪脱氢球菌菌株由于能够将所有氯化乙烯(最著名的是致癌的氯乙烯)脱氯成乙烯的能力而得到了最广泛的研究。OHRB的基因组,特别是专一的OHRB,通常具有多个推定的还原性脱卤酶基因(rdhA),其中大多数尚未鉴定。我们最近对8个新菌株的基因组进行了测序并关闭了基因组,从而增加了可用的D. mccartyi数量。从16到24的NCBI基因组。从所有可用的OHRB基因组中,我们使用先前建立的90%氨基酸成对同一性临界值来识别Ortholog组(OGs),对还原性脱卤素酶基因的预测翻译进行分类。有趣的是,大多数麦卡迪脱卤素酶基因序列一旦被归类为OG,则在迄今测序的所有基因组中均表现出显着的协同性(基因顺序)。没有分类,这个组织是不明显的。高度的一致性表明差异是由于rdhA基因丢失而不是重组引起的。系统发育分析表明,大多数rdhA基因的形态具有与原点Dehalococcoidia漫长的进化历史之前Dehalococcoides和Dehalogenimonas。我们还寻找其他OHRB物种的基因组中的同构证据。不幸的是,此时封闭的Dehalogenimonas基因组太少,无法比较。有部分证据表明限制性脱氢杆菌的基因组存在同义,但是这里需要更多封闭基因组进行确认。有趣的是,我们发现,rdhA基因编码的酶,工业污染物的催化脱卤是唯一rdhA具有最近横向转移的有力证据的基因-至少在本文研究的基因组中。鉴于RdhA序列分类可用于比较分析,我们正在构建一个公共Web服务器(http://RDaseDB.biozone.utoronto.ca)供社区使用,它允许用户添加和分类新序列,并下载完整的还原性脱卤素酶数据库。
更新日期:2020-04-24
中文翻译:

通过基因组组装和直系同源物组分类,了解还原性脱卤化氢酶基因家族的未开发多数的起源和功能。
有机卤化物呼吸细菌(OHRB)表达还原性脱卤素酶,用于能量保存和生长。这些酶中的某些可在厌氧地下环境中催化氯化物和溴化污染物的还原脱卤作用,从而提供有价值的生态系统服务。麦卡迪脱氢球菌菌株由于能够将所有氯化乙烯(最著名的是致癌的氯乙烯)脱氯成乙烯的能力而得到了最广泛的研究。OHRB的基因组,特别是专一的OHRB,通常具有多个推定的还原性脱卤酶基因(rdhA),其中大多数尚未鉴定。我们最近对8个新菌株的基因组进行了测序并关闭了基因组,从而增加了可用的D. mccartyi数量。从16到24的NCBI基因组。从所有可用的OHRB基因组中,我们使用先前建立的90%氨基酸成对同一性临界值来识别Ortholog组(OGs),对还原性脱卤素酶基因的预测翻译进行分类。有趣的是,大多数麦卡迪脱卤素酶基因序列一旦被归类为OG,则在迄今测序的所有基因组中均表现出显着的协同性(基因顺序)。没有分类,这个组织是不明显的。高度的一致性表明差异是由于rdhA基因丢失而不是重组引起的。系统发育分析表明,大多数rdhA基因的形态具有与原点Dehalococcoidia漫长的进化历史之前Dehalococcoides和Dehalogenimonas。我们还寻找其他OHRB物种的基因组中的同构证据。不幸的是,此时封闭的Dehalogenimonas基因组太少,无法比较。有部分证据表明限制性脱氢杆菌的基因组存在同义,但是这里需要更多封闭基因组进行确认。有趣的是,我们发现,rdhA基因编码的酶,工业污染物的催化脱卤是唯一rdhA具有最近横向转移的有力证据的基因-至少在本文研究的基因组中。鉴于RdhA序列分类可用于比较分析,我们正在构建一个公共Web服务器(http://RDaseDB.biozone.utoronto.ca)供社区使用,它允许用户添加和分类新序列,并下载完整的还原性脱卤素酶数据库。











































 京公网安备 11010802027423号
京公网安备 11010802027423号