当前位置:
X-MOL 学术
›
Cancer Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of hub methylated-CpG sites and associated genes in oral squamous cell carcinoma.
Cancer Medicine ( IF 2.9 ) Pub Date : 2020-03-10 , DOI: 10.1002/cam4.2969 Yuxin Dai 1, 2 , Qiaoli Lv 3 , Tingting Qi 4 , Jian Qu 4 , Hongli Ni 1 , Yongkang Liao 5 , Peng Liu 5 , Qiang Qu 1, 6
Cancer Medicine ( IF 2.9 ) Pub Date : 2020-03-10 , DOI: 10.1002/cam4.2969 Yuxin Dai 1, 2 , Qiaoli Lv 3 , Tingting Qi 4 , Jian Qu 4 , Hongli Ni 1 , Yongkang Liao 5 , Peng Liu 5 , Qiang Qu 1, 6
Affiliation
|
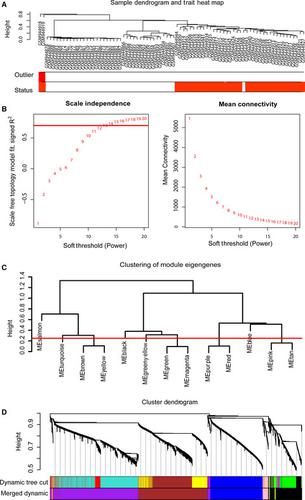
To improve personalized diagnosis and prognosis for oral squamous cell carcinoma (OSCC) by identification of hub methylated-CpG sites and associated genes, weighted gene comethylation network analysis (WGCNA) was performed to examine and identify hub modules and CpG sites correlated with OSCC. Here, WGCNA modeling yielded blue and brown comethylation modules that were significantly associated with OSCC status. Following screening of the differentially expressed genes (DEGs) from gene expression microarrays and differentially methylated-CpG sites (DCGs), integrated multiomics analysis of the DEGs, DCGs, and hub CpG sites from the modules was performed to investigate their correlations. Expression levels of 16 CpG sites-associated genes were negatively correlated with methylation patterns of promoter. Moreover, Kaplan-Meier survival analysis of the hub CpG sites and associated genes was carried out using 2 public databases, MethSurv and GEPIA. Only 5 genes, ACTA1, ACTN2, OSR1, SYNGR1, and ZNF677, had significant overall survival using GEPIA. Hypermethylated-CpG sites ACTN2-cg21376883 and OSR1-cg06509239 were found to be associated with poor survival by MethSurv. Methylation status of specific site and expression levels of associated genes were determined using clinical samples by quantitative methylation-specific PCR and real-time PCR. Pearson's correlation analysis showed that methylation levels of cg06509239 and cg18335068 were negatively related to OSR1 and ZNF677 expression levels, respectively. Our classification schema using multiomics analysis represents a screening framework for identification of hub CpG sites and associated genes.
中文翻译:

口腔鳞状细胞癌中枢甲基化的CpG位点和相关基因的鉴定。
为了通过识别毂甲基化的CpG位点和相关基因来改善口腔鳞状细胞癌(OSCC)的个性化诊断和预后,进行了加权基因共甲基化网络分析(WGCNA)来检查和鉴定与OSCC相关的毂模块和CpG位点。在这里,WGCNA建模产生与OSCC状态显着相关的蓝色和棕色共甲基化模块。在从基因表达微阵列和差异甲基化CpG位点(DCG)筛选差异表达基因(DEG)之后,对模块中的DEG,DCG和集线器CpG位点进行了集成多组学分析,以研究它们之间的相关性。16个CpG位点相关基因的表达水平与启动子的甲基化模式呈负相关。此外,使用2个公共数据库MethSurv和GEPIA对中枢CpG位点和相关基因进行Kaplan-Meier生存分析。使用GEPIA时,只有5个基因ACTAA1,ACTN2,OSR1,SYNGR1和ZNF677具有显着的总体存活率。MethSurv发现超甲基化CpG位点ACTN2-cg21376883和OSR1-cg06509239与存活率低有关。通过定量甲基化特异性PCR和实时PCR,使用临床样品确定特异性位点的甲基化状态和相关基因的表达水平。Pearson的相关分析表明,cg06509239和cg18335068的甲基化水平分别与OSR1和ZNF677表达水平呈负相关。我们使用多组学分析的分类方案代表了用于鉴定中心CpG位点和相关基因的筛选框架。
更新日期:2020-03-10
中文翻译:

口腔鳞状细胞癌中枢甲基化的CpG位点和相关基因的鉴定。
为了通过识别毂甲基化的CpG位点和相关基因来改善口腔鳞状细胞癌(OSCC)的个性化诊断和预后,进行了加权基因共甲基化网络分析(WGCNA)来检查和鉴定与OSCC相关的毂模块和CpG位点。在这里,WGCNA建模产生与OSCC状态显着相关的蓝色和棕色共甲基化模块。在从基因表达微阵列和差异甲基化CpG位点(DCG)筛选差异表达基因(DEG)之后,对模块中的DEG,DCG和集线器CpG位点进行了集成多组学分析,以研究它们之间的相关性。16个CpG位点相关基因的表达水平与启动子的甲基化模式呈负相关。此外,使用2个公共数据库MethSurv和GEPIA对中枢CpG位点和相关基因进行Kaplan-Meier生存分析。使用GEPIA时,只有5个基因ACTAA1,ACTN2,OSR1,SYNGR1和ZNF677具有显着的总体存活率。MethSurv发现超甲基化CpG位点ACTN2-cg21376883和OSR1-cg06509239与存活率低有关。通过定量甲基化特异性PCR和实时PCR,使用临床样品确定特异性位点的甲基化状态和相关基因的表达水平。Pearson的相关分析表明,cg06509239和cg18335068的甲基化水平分别与OSR1和ZNF677表达水平呈负相关。我们使用多组学分析的分类方案代表了用于鉴定中心CpG位点和相关基因的筛选框架。











































 京公网安备 11010802027423号
京公网安备 11010802027423号