npj Digital Medicine ( IF 12.4 ) Pub Date : 2020-03-10 , DOI: 10.1038/s41746-020-0238-2 Shivam Kalra 1, 2 , H R Tizhoosh 2, 3 , Sultaan Shah 1 , Charles Choi 1 , Savvas Damaskinos 1 , Amir Safarpoor 2 , Sobhan Shafiei 2 , Morteza Babaie 2 , Phedias Diamandis 4 , Clinton J V Campbell 5, 6 , Liron Pantanowitz 7
|
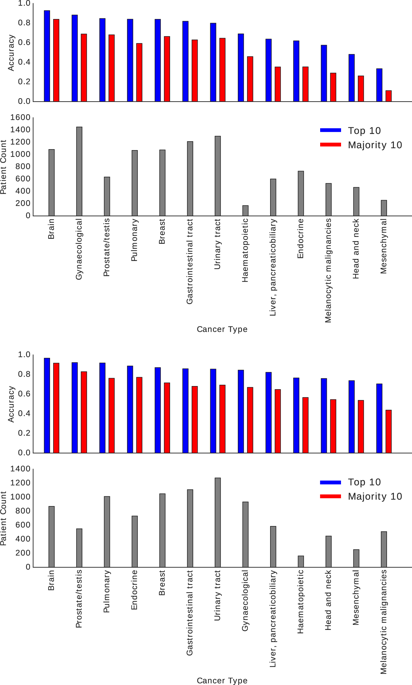
The emergence of digital pathology has opened new horizons for histopathology. Artificial intelligence (AI) algorithms are able to operate on digitized slides to assist pathologists with different tasks. Whereas AI-involving classification and segmentation methods have obvious benefits for image analysis, image search represents a fundamental shift in computational pathology. Matching the pathology of new patients with already diagnosed and curated cases offers pathologists a new approach to improve diagnostic accuracy through visual inspection of similar cases and computational majority vote for consensus building. In this study, we report the results from searching the largest public repository (The Cancer Genome Atlas, TCGA) of whole-slide images from almost 11,000 patients. We successfully indexed and searched almost 30,000 high-resolution digitized slides constituting 16 terabytes of data comprised of 20 million 1000 × 1000 pixels image patches. The TCGA image database covers 25 anatomic sites and contains 32 cancer subtypes. High-performance storage and GPU power were employed for experimentation. The results were assessed with conservative “majority voting” to build consensus for subtype diagnosis through vertical search and demonstrated high accuracy values for both frozen section slides (e.g., bladder urothelial carcinoma 93%, kidney renal clear cell carcinoma 97%, and ovarian serous cystadenocarcinoma 99%) and permanent histopathology slides (e.g., prostate adenocarcinoma 98%, skin cutaneous melanoma 99%, and thymoma 100%). The key finding of this validation study was that computational consensus appears to be possible for rendering diagnoses if a sufficiently large number of searchable cases are available for each cancer subtype.
中文翻译:

通过人工智能搜索档案组织病理学图像达成泛癌诊断共识
数字病理学的出现为组织病理学开辟了新的视野。人工智能 (AI) 算法能够在数字化载玻片上运行,以协助病理学家完成不同的任务。尽管涉及人工智能的分类和分割方法对于图像分析具有明显的好处,但图像搜索代表了计算病理学的根本转变。将新患者的病理与已诊断和策划的病例进行匹配,为病理学家提供了一种新方法,通过对类似病例的目视检查和计算多数投票建立共识来提高诊断准确性。在这项研究中,我们报告了搜索最大公共存储库(癌症基因组图谱,TCGA)的结果,其中包含来自近 11,000 名患者的全幻灯片图像。我们成功地索引和搜索了近 30,000 张高分辨率数字化幻灯片,这些幻灯片构成了 16 TB 的数据,其中包含 2000 万个 1000 × 1000 像素图像块。 TCGA 图像数据库涵盖 25 个解剖部位,包含 32 种癌症亚型。实验采用了高性能存储和 GPU 能力。结果通过保守的“多数投票”进行评估,通过垂直搜索建立亚型诊断共识,并证明了两种冰冻切片切片的高精度值(例如,膀胱尿路上皮癌 93%、肾肾透明细胞癌 97% 和卵巢浆液性囊腺癌) 99%)和永久性组织病理学切片(例如,前列腺腺癌 98%、皮肤黑色素瘤 99% 和胸腺瘤 100%)。这项验证研究的主要发现是,如果每种癌症亚型都有足够多的可搜索病例,那么计算共识似乎可以用于诊断。











































 京公网安备 11010802027423号
京公网安备 11010802027423号