当前位置:
X-MOL 学术
›
Anal. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A fluorescence-positioned hybridization chain reaction system for sensitive detection of Salmonella in milk
Analytical Methods ( IF 2.7 ) Pub Date : 2020-03-07 , DOI: 10.1039/d0ay00184h Guotai Yang 1, 2, 3, 4 , Shuang Yu 1, 2, 3, 4 , Yang Liu 5, 6, 7 , Jin Huang 1, 2, 3, 4 , Qianying Li 1, 2, 3, 4 , Zoraida P. Aguilar 8, 9, 10 , Hengyi Xu 1, 2, 3, 4
Analytical Methods ( IF 2.7 ) Pub Date : 2020-03-07 , DOI: 10.1039/d0ay00184h Guotai Yang 1, 2, 3, 4 , Shuang Yu 1, 2, 3, 4 , Yang Liu 5, 6, 7 , Jin Huang 1, 2, 3, 4 , Qianying Li 1, 2, 3, 4 , Zoraida P. Aguilar 8, 9, 10 , Hengyi Xu 1, 2, 3, 4
Affiliation
|
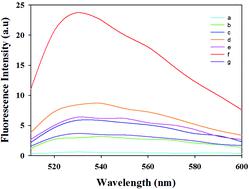
In this study, a fluorescence-positioned hybridization chain reaction (HCR) system for the detection of single-stranded DNA (ssDNA) specific to Salmonella was developed. Target ssDNA captured by magnetic beads was used to trigger the HCR, forming multiple double-stranded DNAs (dsDNAs). SYBR Green I was applied to intercalate with the minor groove of the dsDNA forming MBs + dsDNA + SYBR Green I. Magnetic separation was used to isolate the MBs + dsDNA + SYBR Green I from the excess reagents before recording the fluorescence intensity (FI). This fluorescence-positioned HCR method for the quantitative detection of Salmonella showed a limit of detection (LOD) as low as 4.2 × 101 CFU mL−1 in 0.01 M phosphate-buffered saline (PBS). When the method was applied in spiked milk samples, the LOD was recorded at 4.2 × 102 CFU mL−1. The fluorescence-positioned HCR method provided a modification-free and fluorescence-positioned strategy for sensitive detection of target DNA which holds promise for sensitive and selective bacterial detection.
中文翻译:

用于牛奶中沙门氏菌灵敏检测的荧光定位杂交链反应系统
在这项研究中,开发了一种荧光定位杂交链反应(HCR)系统,用于检测沙门氏菌特异的单链DNA(ssDNA)。磁珠捕获的目标ssDNA用于触发HCR,形成多个双链DNA(dsDNA)。将SYBR Green I插入到dsDNA的小沟中,形成MBs + dsDNA + SYBR GreenI。在记录荧光强度(FI)之前,用磁分离法从多余的试剂中分离出MBs + dsDNA + SYBR GreenI。此荧光定位的HCR方法用于沙门氏菌的定量检测,检测限(LOD)低至4.2×10 1 CFU mL -1加入0.01 M磷酸盐缓冲液(PBS)中。当该方法应用于加标牛奶样品时,LOD记录为4.2×10 2 CFU mL -1。荧光定位的HCR方法为目标DNA的灵敏检测提供了无修饰和荧光定位的策略,为敏感和选择性的细菌检测提供了希望。
更新日期:2020-04-24
中文翻译:

用于牛奶中沙门氏菌灵敏检测的荧光定位杂交链反应系统
在这项研究中,开发了一种荧光定位杂交链反应(HCR)系统,用于检测沙门氏菌特异的单链DNA(ssDNA)。磁珠捕获的目标ssDNA用于触发HCR,形成多个双链DNA(dsDNA)。将SYBR Green I插入到dsDNA的小沟中,形成MBs + dsDNA + SYBR GreenI。在记录荧光强度(FI)之前,用磁分离法从多余的试剂中分离出MBs + dsDNA + SYBR GreenI。此荧光定位的HCR方法用于沙门氏菌的定量检测,检测限(LOD)低至4.2×10 1 CFU mL -1加入0.01 M磷酸盐缓冲液(PBS)中。当该方法应用于加标牛奶样品时,LOD记录为4.2×10 2 CFU mL -1。荧光定位的HCR方法为目标DNA的灵敏检测提供了无修饰和荧光定位的策略,为敏感和选择性的细菌检测提供了希望。











































 京公网安备 11010802027423号
京公网安备 11010802027423号