当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Agonist-Specific Conformations of the M2 Muscarinic Acetylcholine Receptor Assessed by Molecular Dynamics.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-03-04 , DOI: 10.1021/acs.jcim.0c00041 Alena Randáková 1 , Dominik Nelic 1 , Vladimír Doležal 1 , Esam E El-Fakahany 2 , John Boulos 3 , Jan Jakubík 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-03-04 , DOI: 10.1021/acs.jcim.0c00041 Alena Randáková 1 , Dominik Nelic 1 , Vladimír Doležal 1 , Esam E El-Fakahany 2 , John Boulos 3 , Jan Jakubík 1
Affiliation
|
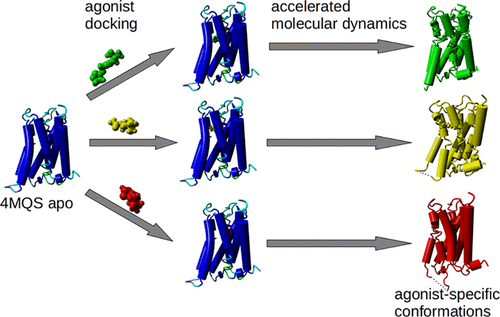
Binding of muscarinic ligands, both antagonists and agonists, and their effects on the conformation of the M2 acetylcholine receptor were modeled in silico and compared to experimental data. After docking of antagonists to the M2 receptor in an inactive conformation (3UON, 5ZK3, 5ZKB, or 5ZKB) and agonists in an active conformation (4MQS), 100 ns of conventional molecular dynamics (MD) followed by 500 ns of accelerated MD was run. Conventional MD revealed ligand-specific interactions with the receptor. Antagonists stabilized the receptor in an inactive conformation during accelerated MD. The receptor in complex with various agonists attained different conformations specific to individual agonists. The magnitude of the TM6 movement correlated with agonist efficacy at the non-preferential Gs pathway. The shape of the intracellular opening where the receptor interacts with a G-protein was different for the classical agonist carbachol, super-agonist iperoxo, and Gi/o-biased partial agonists JR-6 and JR-7, being compatible with experimentally observed agonist bias at the G-protein level. Moreover, a wash-resistant binding of the unique agonist xanomeline associated with interactions with membrane lipids was formed during accelerated MD. Thus, accelerated MD is suitable for modeling of ligand-specific receptor binding and receptor conformations that is essential for the design of experiments aimed at identification of the secondary binding sites and understanding molecular mechanisms underlying receptor activation.
中文翻译:

M2毒蕈碱型乙酰胆碱受体的激动剂特异性构象,通过分子动力学评估。
在计算机模拟了毒蕈碱配体(拮抗剂和激动剂)的结合及其对M2乙酰胆碱受体构象的影响,并与实验数据进行了比较。拮抗剂以无活性构象(3UON,5ZK3、5ZKB或5ZKB)和激动剂以活性构象(4MQS)对接后,运行100 ns的常规分子动力学(MD),然后运行500 ns的加速MD 。常规MD揭示了与受体的配体特异性相互作用。拮抗剂在加速的MD期间将受体稳定在非活性构象中。与各种激动剂复合的受体获得了对单个激动剂特异的不同构象。TM6运动的幅度与非优先Gs途径的激动剂功效相关。受体与G蛋白相互作用的细胞内开口的形状对于经典激动剂卡巴胆碱,超级激动剂iperoxo和Gi / o偏向部分激动剂JR-6和JR-7是不同的,与实验观察到的激动剂相容在G蛋白水平上存在偏见。而且,在加速的MD期间形成了独特的激动剂黄嘌呤的耐洗涤结合与与膜脂质的相互作用有关。因此,加速的MD适用于建模配体特异性受体结合和受体构象,这对于设计旨在鉴定次级结合位点和理解受体激活基础的分子机制的实验是必不可少的。和Gi / o偏向的部分激动剂JR-6和JR-7,与在G蛋白水平上实验观察到的激动剂偏倚兼容。而且,在加速的MD期间形成了独特的激动剂黄嘌呤的耐洗涤结合与与膜脂质的相互作用有关。因此,加速的MD适用于建模配体特异性受体结合和受体构象,这对于设计旨在鉴定次级结合位点和理解受体激活基础的分子机制的实验是必不可少的。和Gi / o偏向的部分激动剂JR-6和JR-7,与在G蛋白水平上实验观察到的激动剂偏倚兼容。而且,在加速的MD期间形成了独特的激动剂黄嘌呤的耐洗涤结合与与膜脂质的相互作用有关。因此,加速的MD适用于建模配体特异性受体结合和受体构象,这对于设计旨在鉴定次级结合位点和理解受体激活基础的分子机制的实验是必不可少的。
更新日期:2020-03-04
中文翻译:

M2毒蕈碱型乙酰胆碱受体的激动剂特异性构象,通过分子动力学评估。
在计算机模拟了毒蕈碱配体(拮抗剂和激动剂)的结合及其对M2乙酰胆碱受体构象的影响,并与实验数据进行了比较。拮抗剂以无活性构象(3UON,5ZK3、5ZKB或5ZKB)和激动剂以活性构象(4MQS)对接后,运行100 ns的常规分子动力学(MD),然后运行500 ns的加速MD 。常规MD揭示了与受体的配体特异性相互作用。拮抗剂在加速的MD期间将受体稳定在非活性构象中。与各种激动剂复合的受体获得了对单个激动剂特异的不同构象。TM6运动的幅度与非优先Gs途径的激动剂功效相关。受体与G蛋白相互作用的细胞内开口的形状对于经典激动剂卡巴胆碱,超级激动剂iperoxo和Gi / o偏向部分激动剂JR-6和JR-7是不同的,与实验观察到的激动剂相容在G蛋白水平上存在偏见。而且,在加速的MD期间形成了独特的激动剂黄嘌呤的耐洗涤结合与与膜脂质的相互作用有关。因此,加速的MD适用于建模配体特异性受体结合和受体构象,这对于设计旨在鉴定次级结合位点和理解受体激活基础的分子机制的实验是必不可少的。和Gi / o偏向的部分激动剂JR-6和JR-7,与在G蛋白水平上实验观察到的激动剂偏倚兼容。而且,在加速的MD期间形成了独特的激动剂黄嘌呤的耐洗涤结合与与膜脂质的相互作用有关。因此,加速的MD适用于建模配体特异性受体结合和受体构象,这对于设计旨在鉴定次级结合位点和理解受体激活基础的分子机制的实验是必不可少的。和Gi / o偏向的部分激动剂JR-6和JR-7,与在G蛋白水平上实验观察到的激动剂偏倚兼容。而且,在加速的MD期间形成了独特的激动剂黄嘌呤的耐洗涤结合与与膜脂质的相互作用有关。因此,加速的MD适用于建模配体特异性受体结合和受体构象,这对于设计旨在鉴定次级结合位点和理解受体激活基础的分子机制的实验是必不可少的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号