当前位置:
X-MOL 学术
›
ACS Comb. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Critical Evaluation of Photo-cross-linking Parameters for the Implementation of Efficient DNA-Encoded Chemical Library Selections.
ACS Combinatorial Science Pub Date : 2020-03-06 , DOI: 10.1021/acscombsci.0c00023 Alessandro Sannino 1 , Adrián Gironda-Martínez 1 , Émile M D Gorre 1 , Luca Prati 1 , Jacopo Piazzi 1 , Jörg Scheuermann 2 , Dario Neri 2 , Etienne J Donckele 1 , Florent Samain 1
ACS Combinatorial Science Pub Date : 2020-03-06 , DOI: 10.1021/acscombsci.0c00023 Alessandro Sannino 1 , Adrián Gironda-Martínez 1 , Émile M D Gorre 1 , Luca Prati 1 , Jacopo Piazzi 1 , Jörg Scheuermann 2 , Dario Neri 2 , Etienne J Donckele 1 , Florent Samain 1
Affiliation
|
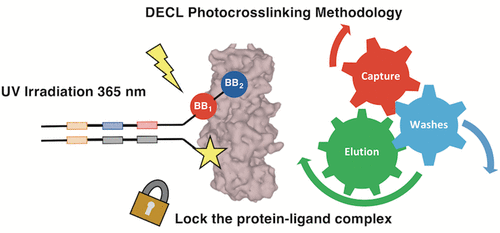
The growing importance of DNA-encoded chemical libraries (DECLs) as tools for the discovery of protein binders has sparked an interest for the development of efficient screening methodologies, capable of discriminating between high- and medium-affinity ligands. Here, we present a systematic investigation of selection methodologies, featuring a library displayed on single-stranded DNA, which could be hybridized to a complementary oligonucleotide carrying a diazirine photoreactive group. Model experiments, performed using ligands of different affinity to carbonic anhydrase IX, revealed a recovery of preferential binders up to 10%, which was mainly limited by the highly reactive nature of carbene intermediates generated during the photo-cross-linking process. Ligands featuring acetazolamide or p-phenylsulfonamide exhibited a higher recovery compared to their counterparts based on 3-sulfamoyl benzoic acid, which had a lower affinity toward the target. A systematic evaluation of experimental parameters revealed conditions that were ideally suited for library screening, which were used for the screening of a combinatorial DECL library, featuring 669 240 combinations of two sets of building blocks. Compared to conventional affinity capture procedures on protein immobilized on solid supports, photo-cross-linking provided a better discrimination of low-affinity CAIX ligands over the background signal and therefore can be used as a tandem methodology with the affinity capture procedures.
中文翻译:

关键评估的光交联参数,以实现有效的DNA编码化学文库选择。
DNA编码化学文库(DECL)作为发现蛋白结合剂的工具的重要性与日俱增,引起了人们对开发能够区分高亲和力和中亲和力配体的高效筛选方法学的兴趣。在这里,我们介绍选择方法的系统研究,其特征是展示在单链DNA上的文库可以与带有diazirine光反应性基团的互补寡核苷酸杂交。使用对碳酸酐酶IX具有不同亲和力的配体进行的模型实验显示,最多可回收10%的优先结合剂,这主要受光交联过程中生成的卡宾中间体的高反应性限制。与基于3-氨磺酰基苯甲酸的对应物相比,具有乙酰唑胺或对苯磺酰胺的配体表现出更高的回收率,后者对靶标的亲和力较低。对实验参数的系统评估揭示了非常适合文库筛选的条件,该条件用于筛选组合DECL文库,其特征是两组构建基的669 240个组合。与固定在固体支持物上的蛋白质的常规亲和力捕获程序相比,光交联可更好地区分背景信号上的低亲和力CAIX配体,因此可与亲和力捕获程序一起用作串联方法。对实验参数的系统评估揭示了非常适合文库筛选的条件,这些条件用于筛选组合DECL文库,其中包含两组构建基的669240个组合。与固定在固体支持物上的蛋白质的常规亲和力捕获程序相比,光交联可更好地区分背景信号上的低亲和力CAIX配体,因此可与亲和力捕获程序一起用作串联方法。对实验参数的系统评估揭示了非常适合文库筛选的条件,这些条件用于筛选组合DECL文库,其中包含两组构建基的669240个组合。与固定在固体支持物上的蛋白质的常规亲和力捕获程序相比,光交联可更好地区分背景信号上的低亲和力CAIX配体,因此可与亲和力捕获程序一起用作串联方法。
更新日期:2020-02-28
中文翻译:

关键评估的光交联参数,以实现有效的DNA编码化学文库选择。
DNA编码化学文库(DECL)作为发现蛋白结合剂的工具的重要性与日俱增,引起了人们对开发能够区分高亲和力和中亲和力配体的高效筛选方法学的兴趣。在这里,我们介绍选择方法的系统研究,其特征是展示在单链DNA上的文库可以与带有diazirine光反应性基团的互补寡核苷酸杂交。使用对碳酸酐酶IX具有不同亲和力的配体进行的模型实验显示,最多可回收10%的优先结合剂,这主要受光交联过程中生成的卡宾中间体的高反应性限制。与基于3-氨磺酰基苯甲酸的对应物相比,具有乙酰唑胺或对苯磺酰胺的配体表现出更高的回收率,后者对靶标的亲和力较低。对实验参数的系统评估揭示了非常适合文库筛选的条件,该条件用于筛选组合DECL文库,其特征是两组构建基的669 240个组合。与固定在固体支持物上的蛋白质的常规亲和力捕获程序相比,光交联可更好地区分背景信号上的低亲和力CAIX配体,因此可与亲和力捕获程序一起用作串联方法。对实验参数的系统评估揭示了非常适合文库筛选的条件,这些条件用于筛选组合DECL文库,其中包含两组构建基的669240个组合。与固定在固体支持物上的蛋白质的常规亲和力捕获程序相比,光交联可更好地区分背景信号上的低亲和力CAIX配体,因此可与亲和力捕获程序一起用作串联方法。对实验参数的系统评估揭示了非常适合文库筛选的条件,这些条件用于筛选组合DECL文库,其中包含两组构建基的669240个组合。与固定在固体支持物上的蛋白质的常规亲和力捕获程序相比,光交联可更好地区分背景信号上的低亲和力CAIX配体,因此可与亲和力捕获程序一起用作串联方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号