当前位置:
X-MOL 学术
›
Genes Brain Behav.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Genome-wide association study of word reading: Overlap with risk genes for neurodevelopmental disorders.
Genes, Brain and Behavior ( IF 2.4 ) Pub Date : 2020-02-27 , DOI: 10.1111/gbb.12648 Kaitlyn M Price 1, 2, 3 , Karen G Wigg 1 , Yu Feng 1 , Kirsten Blokland 2 , Margaret Wilkinson 2 , Gengming He 4 , Elizabeth N Kerr 5, 6 , Tasha-Cate Carter 2, 7 , Sharon L Guger 5 , Maureen W Lovett 2, 6 , Lisa J Strug 4, 8 , Cathy L Barr 1, 2, 3
Genes, Brain and Behavior ( IF 2.4 ) Pub Date : 2020-02-27 , DOI: 10.1111/gbb.12648 Kaitlyn M Price 1, 2, 3 , Karen G Wigg 1 , Yu Feng 1 , Kirsten Blokland 2 , Margaret Wilkinson 2 , Gengming He 4 , Elizabeth N Kerr 5, 6 , Tasha-Cate Carter 2, 7 , Sharon L Guger 5 , Maureen W Lovett 2, 6 , Lisa J Strug 4, 8 , Cathy L Barr 1, 2, 3
Affiliation
|
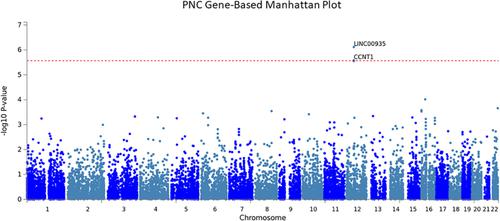
Reading disabilities (RD) are the most common neurocognitive disorder, affecting 5% to 17% of children in North America. These children often have comorbid neurodevelopmental/psychiatric disorders, such as attention deficit/hyperactivity disorder (ADHD). The genetics of RD and their overlap with other disorders is incompletely understood. To contribute to this, we performed a genome‐wide association study (GWAS) for word reading. Then, using summary statistics from neurodevelopmental/psychiatric disorders, we computed polygenic risk scores (PRS) and used them to predict reading ability in our samples. This enabled us to test the shared aetiology between RD and other disorders. The GWAS consisted of 5.3 million single nucleotide polymorphisms (SNPs) and two samples; a family‐based sample recruited for reading difficulties in Toronto (n = 624) and a population‐based sample recruited in Philadelphia [Philadelphia Neurodevelopmental Cohort (PNC)] (n = 4430). The Toronto sample SNP‐based analysis identified suggestive SNPs (P ~ 5 × 10−7) in the ARHGAP23 gene, which is implicated in neuronal migration/axon pathfinding. The PNC gene‐based analysis identified significant associations (P < 2.72 × 10−6) for LINC00935 and CCNT1 , located in the region of the KANSL2/CCNT1/LINC00935/SNORA2B/SNORA34/MIR4701/ADCY6 genes on chromosome 12q, with near significant SNP‐based analysis. PRS identified significant overlap between word reading and intelligence (R 2 = 0.18, P = 7.25 × 10−181), word reading and educational attainment (R 2 = 0.07, P = 4.91 × 10−48) and word reading and ADHD (R 2 = 0.02, P = 8.70 × 10−6; threshold for significance = 7.14 × 10−3). Overlap was also found between RD and autism spectrum disorder (ASD) as top‐ranked genes were previously implicated in autism by rare and copy number variant analyses. These findings support shared risk between word reading, cognitive measures, educational outcomes and neurodevelopmental disorders, including ASD.
中文翻译:

单词阅读的全基因组关联研究:与神经发育障碍的风险基因重叠。
阅读障碍(RD)是最常见的神经认知障碍,在北美影响5%至17%的儿童。这些儿童通常患有合并症的神经发育/精神疾病,例如注意力不足/多动症(ADHD)。RD的遗传学及其与其他疾病的重叠尚不完全清楚。为此,我们进行了全基因组关联研究(GWAS)以进行单词阅读。然后,使用来自神经发育/精神疾病的摘要统计数据,我们计算了多基因风险评分(PRS),并使用它们来预测样本中的阅读能力。这使我们能够检验RD与其他疾病之间的共同病因。GWAS由530万个单核苷酸多态性(SNP)和两个样本组成。一个在多伦多因阅读困难而招募的家庭样本(624例)和在费城(Philadelphia Neurodevelopmental Cohort,宾夕法尼亚州)招募了一个基于人群的样本(n = 4430)。多伦多样本基于SNP的分析确定了提示性SNP(P〜5×10 -7)在ARHGAP23基因,这是牵涉于神经元迁移/轴突寻路。基于PNC基因的分析确定了LINC00935和CCNT1的显着关联(P <2.72×10 -6),它们位于12q染色体上的KANSL2 / CCNT1 / LINC00935 / SNORA2B / SNORA34 / MIR4701 / ADCY6基因区域基于SNP的分析。PRS发现单词阅读和智力(R 2 = 0.18,P = 7.25×10 -181),单词阅读和教育程度(R 2 = 0.07,P = 4.91×10 -48)以及单词阅读和ADHD(R 2 = 0.02,P = 8.70×10 -6;有意义的阈值= 7.14×10 -3)。在RD和自闭症谱系障碍(ASD)之间也发现了重叠,因为先前通过稀有和拷贝数变异分析将排名靠前的基因牵涉到自闭症中。这些发现支持单词阅读,认知措施,教育成果和神经发育障碍(包括ASD)之间的共同风险。
更新日期:2020-02-27
中文翻译:

单词阅读的全基因组关联研究:与神经发育障碍的风险基因重叠。
阅读障碍(RD)是最常见的神经认知障碍,在北美影响5%至17%的儿童。这些儿童通常患有合并症的神经发育/精神疾病,例如注意力不足/多动症(ADHD)。RD的遗传学及其与其他疾病的重叠尚不完全清楚。为此,我们进行了全基因组关联研究(GWAS)以进行单词阅读。然后,使用来自神经发育/精神疾病的摘要统计数据,我们计算了多基因风险评分(PRS),并使用它们来预测样本中的阅读能力。这使我们能够检验RD与其他疾病之间的共同病因。GWAS由530万个单核苷酸多态性(SNP)和两个样本组成。一个在多伦多因阅读困难而招募的家庭样本(624例)和在费城(Philadelphia Neurodevelopmental Cohort,宾夕法尼亚州)招募了一个基于人群的样本(n = 4430)。多伦多样本基于SNP的分析确定了提示性SNP(P〜5×10 -7)在ARHGAP23基因,这是牵涉于神经元迁移/轴突寻路。基于PNC基因的分析确定了LINC00935和CCNT1的显着关联(P <2.72×10 -6),它们位于12q染色体上的KANSL2 / CCNT1 / LINC00935 / SNORA2B / SNORA34 / MIR4701 / ADCY6基因区域基于SNP的分析。PRS发现单词阅读和智力(R 2 = 0.18,P = 7.25×10 -181),单词阅读和教育程度(R 2 = 0.07,P = 4.91×10 -48)以及单词阅读和ADHD(R 2 = 0.02,P = 8.70×10 -6;有意义的阈值= 7.14×10 -3)。在RD和自闭症谱系障碍(ASD)之间也发现了重叠,因为先前通过稀有和拷贝数变异分析将排名靠前的基因牵涉到自闭症中。这些发现支持单词阅读,认知措施,教育成果和神经发育障碍(包括ASD)之间的共同风险。









































 京公网安备 11010802027423号
京公网安备 11010802027423号